Transcript Levels of Aldo-Keto Reductase Family 1 Subfamily C (AKR1C) Are Increased in Prostate Tissue of Patients with Type 2 Diabetes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Taylor Et Al REVISED MS28926-1.Pdf
1 The effect of micronutrient supplementation on growth and hepatic 2 metabolism in diploid and triploid Atlantic salmon (Salmo salar) parr 3 fed a low marine ingredient diet 4 5 John F. Taylor a*, Luisa M. Vera a, Christian De Santis a, Erik-Jan Lock b, Marit Espe b, 6 Kaja H. Skjærven b, Daniel Leeming c, Jorge del Pozo d, Jose Mota-Velasco e, Herve 7 Migaud a, Kristin Hamre b, Douglas R. Tocher a 8 9 a Institute of Aquaculture, University of Stirling, Stirling, FK9 4LA, UK 10 b Institute of Marine Research, PO box 1870 Nordnes, 5817 Bergen, Norway 11 c BioMar Ltd., Grangemouth, FK3 8UL, UK 12 d The Royal (Dick) School of Veterinary Studies, Edinburgh, EH25 9RG, UK 13 e Hendrix Genetics, Landcatch Natural Selection Ltd., Lochgilphead, PA31 8PE, UK 14 15 Running Title: Dietary micronutrient supplementation in Atlantic salmon 16 17 ms. has 31 pg.s, 4 figures, 9 tables, 4 suppl. files 18 19 Corresponding Author: 20 Dr John F. Taylor 21 Institute of Aquaculture, University of Stirling, Stirling, FK9 4LA, UK 22 Tel: +44-01786 467929 ; Fax: +44-01768 472133 23 [email protected] 24 Accepted refereed manuscript of: Taylor JF, Vera LM, De Santis C, Lock E, Espe M, Skjaerven KH, Leeming D, Del Pozo J, Mota-Velasco J, Migaud H, Hamre K & Tocher DR (2019) The effect of micronutrient supplementation on growth and hepatic metabolism in diploid and triploid Atlantic salmon (Salmo salar) parr fed a low marine ingredient diet. Comparative Biochemistry and Physiology. Part B, Biochemistry and Molecular Biology, 227, pp. -

The Identification of Human Aldo-Keto Reductase AKR7A2 As a Novel
Li et al. Cellular & Molecular Biology Letters (2016) 21:25 Cellular & Molecular DOI 10.1186/s11658-016-0026-9 Biology Letters SHORTCOMMUNICATION Open Access The identification of human aldo-keto reductase AKR7A2 as a novel cytoglobin- binding partner Xin Li, Shanshan Zou, Zhen Li, Gaotai Cai, Bohong Chen, Ping Wang and Wenqi Dong* * Correspondence: [email protected] Abstract Department of Biopharmaceutics, School of Laboratory Medicine and Cytoglobin (CYGB), a member of the globin family, is thought to protect cells from Biotechnology, Southern Medical reactive oxygen and nitrogen species and deal with hypoxic conditions and University, 1838 North Guangzhou oxidative stress. However, its molecular mechanisms of action are not clearly Avenue, Guangzhou 510515, China understood. Through immunoprecipitation combined with a two-dimensional electrophoresis–mass spectrometry assay, we identified a CYGB interactor: aldo-keto reductase family 7 member A2 (AKR7A2). The interaction was further confirmed using yeast two-hybrid and co-immunoprecipitation assays. Our results show that AKR7A2 physically interacts with CYGB. Keywords: CYGB, AKR7A2, Protein-protein interactions, Yeast two-hybrid assay, Co-immunoprecipitation, 2-DE, Oxidative stress Introduction Cytoglobin (CYGB), which is a member of the globin family, was discovered more than a decade ago in a proteomic screen of fibrotic liver [1]. It was originally named STAP (stellate activating protein). Human CYGB is a 190-amino acid, 21-kDa protein [2], encoded by a single copy gene mapped at the 17q25.3 chromosomal segment [3]. It has a compact helical conformation, giving it the ability to bind to heme, which allows reversible binding of gaseous, diatomic molecules, including oxygen (O2), nitric oxide (NO) and carbon monoxide (CO), just like hemoglobin (Hb), myoglobin (Mb) and neuroglobin (Ngb) [4]. -

Gene-Expression Signature Regulated by the KEAP1-NRF2-CUL3 Axis Is Associated with a Poor Prognosis in Head and Neck Squamous Cell Cancer Akhileshwar Namani1†, Md
Namani et al. BMC Cancer (2018) 18:46 DOI 10.1186/s12885-017-3907-z RESEARCH ARTICLE Open Access Gene-expression signature regulated by the KEAP1-NRF2-CUL3 axis is associated with a poor prognosis in head and neck squamous cell cancer Akhileshwar Namani1†, Md. Matiur Rahaman2†, Ming Chen2* and Xiuwen Tang1* Abstract Background: NRF2 is the key regulator of oxidative stress in normal cells and aberrant expression of the NRF2 pathway due to genetic alterations in the KEAP1 (Kelch-like ECH-associated protein 1)-NRF2 (nuclear factor erythroid 2 like 2)-CUL3 (cullin 3) axis leads to tumorigenesis and drug resistance in many cancers including head and neck squamous cell cancer (HNSCC). The main goal of this study was to identify specific genes regulated by the KEAP1-NRF2-CUL3 axis in HNSCC patients, to assess the prognostic value of this gene signature in different cohorts, and to reveal potential biomarkers. Methods: RNA-Seq V2 level 3 data from 279 tumor samples along with 37 adjacent normal samples from patients enrolled in the The Cancer Genome Atlas (TCGA)-HNSCC study were used to identify upregulated genes using two methods (altered KEAP1-NRF2-CUL3 versus normal, and altered KEAP1-NRF2-CUL3 versus wild-type). We then used a new approach to identify the combined gene signature by integrating both datasets and subsequently tested this signature in 4 independent HNSCC datasets to assess its prognostic value. In addition, functional annotation using the DAVID v6.8 database and protein-protein interaction (PPI) analysis using the STRING v10 databasewereperformedonthesignature. Results: A signature composed of a subset of 17 genes regulated by the KEAP1-NRF2-CUL3 axis was identified by overlapping both the upregulated genes of altered versus normal (251 genes) and altered versus wild-type (25 genes) datasets. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Phosphine Stabilizers for Oxidoreductase Enzymes
Europäisches Patentamt *EP001181356B1* (19) European Patent Office Office européen des brevets (11) EP 1 181 356 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.7: C12N 9/02, C12P 7/00, of the grant of the patent: C12P 13/02, C12P 1/00 07.12.2005 Bulletin 2005/49 (86) International application number: (21) Application number: 00917839.3 PCT/US2000/006300 (22) Date of filing: 10.03.2000 (87) International publication number: WO 2000/053731 (14.09.2000 Gazette 2000/37) (54) Phosphine stabilizers for oxidoreductase enzymes Phosphine Stabilisatoren für oxidoreduktase Enzymen Phosphines stabilisateurs des enzymes ayant une activité comme oxidoreducase (84) Designated Contracting States: (56) References cited: DE FR GB NL US-A- 5 777 008 (30) Priority: 11.03.1999 US 123833 P • ABRIL O ET AL.: "Hybrid organometallic/enzymatic catalyst systems: (43) Date of publication of application: Regeneration of NADH using dihydrogen" 27.02.2002 Bulletin 2002/09 JOURNAL OF THE AMERICAN CHEMICAL SOCIETY., vol. 104, no. 6, 1982, pages 1552-1554, (60) Divisional application: XP002148357 DC US cited in the application 05021016.0 • BHADURI S ET AL: "Coupling of catalysis by carbonyl clusters and dehydrigenases: (73) Proprietor: EASTMAN CHEMICAL COMPANY Redution of pyruvate to L-lactate by dihydrogen" Kingsport, TN 37660 (US) JOURNAL OF THE AMERICAN CHEMICAL SOCIETY., vol. 120, no. 49, 11 October 1998 (72) Inventors: (1998-10-11), pages 12127-12128, XP002148358 • HEMBRE, Robert, T. DC US cited in the application Johnson City, TN 37601 (US) • OTSUKA K: "Regeneration of NADH and ketone • WAGENKNECHT, Paul, S. hydrogenation by hydrogen with the San Jose, CA 95129 (US) combination of hydrogenase and alcohol • PENNEY, Jonathan, M. -

Functional Role of Pentose Phosphate Pathway and Glutamine in Cancer Cell
Functional Role of Pentose Phosphate Pathway and Glutamine in Cancer Cell Ibrahim Halil Polat ADVERTIMENT. La consulta d’aquesta tesi queda condicionada a l’acceptació de les següents condicions d'ús: La difusió d’aquesta tesi per mitjà del servei TDX (www.tdx.cat) i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intel·lectual únicament per a usos privats emmarcats en activitats d’investigació i docència. No s’autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d’un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s’autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR (www.tdx.cat) y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. No se autoriza su reproducción con finalidades de lucro ni su difusión y puesta a disposición desde un sitio ajeno al servicio TDR o al Repositorio Digital de la UB. -

Vitamin K1 Prevents Diabetic Cataract by Inhibiting Lens Aldose Reductase 2 (ALR2) Activity Received: 21 May 2019 R
www.nature.com/scientificreports OPEN Vitamin K1 prevents diabetic cataract by inhibiting lens aldose reductase 2 (ALR2) activity Received: 21 May 2019 R. Thiagarajan1,5, M. K. N. Sai Varsha2, V. Srinivasan3, R. Ravichandran4 & K. Saraboji1 Accepted: 24 September 2019 This study investigated the potential of vitamin K1 as a novel lens aldose reductase inhibitor in a Published: xx xx xxxx streptozotocin-induced diabetic cataract model. A single, intraperitoneal injection of streptozotocin (STZ) (35 mg/kg) resulted in hyperglycemia, activation of lens aldose reductase 2 (ALR2) and accumulation of sorbitol in eye lens which could have contributed to diabetic cataract formation. However, when diabetic rats were treated with vitamin K1 (5 mg/kg, sc, twice a week) it resulted in lowering of blood glucose and inhibition of lens aldose reductase activity because of which there was a corresponding decrease in lens sorbitol accumulation. These results suggest that vitamin K1 is a potent inhibitor of lens aldose reductase enzyme and we made an attempt to understand the nature of this inhibition using crude lens homogenate as well as recombinant human aldose reductase enzyme. Our results from protein docking and spectrofuorimetric analyses clearly show that vitamin K1 is a potent inhibitor of ALR2 and this inhibition is primarily mediated by the blockage of DL-glyceraldehyde binding to ALR2. At the same time docking also suggests that vitamin K1 overlaps at the NADPH binding site of ALR2, which probably shows that vitamin K1 could possibly bind both these sites in the enzyme. Another deduction that we can derive from the experiments performed with pure protein is that ALR2 has three levels of afnity, frst for NADPH, second for vitamin K1 and third for the substrate DL-glyceraldehyde. -

Clostridium Difficile Exploits a Host Metabolite Produced During Toxin-Mediated Infection
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.14.426744; this version posted January 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Clostridium difficile exploits a host metabolite produced during toxin-mediated infection 2 3 Kali M. Pruss and Justin L. Sonnenburg* 4 5 Department of Microbiology & Immunology, Stanford University School of Medicine, Stanford 6 CA, USA 94305 7 *Corresponding author address: [email protected] 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 bioRxiv preprint doi: https://doi.org/10.1101/2021.01.14.426744; this version posted January 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 23 Several enteric pathogens can gain specific metabolic advantages over other members of 24 the microbiota by inducing host pathology and inflammation. The pathogen Clostridium 25 difficile (Cd) is responsible for a toxin-mediated colitis that causes 15,000 deaths in the U.S. 26 yearly1, yet the molecular mechanisms by which Cd benefits from toxin-induced colitis 27 remain understudied. Up to 21% of healthy adults are asymptomatic carriers of toxigenic 28 Cd2, indicating that Cd can persist as part of a healthy microbiota; antibiotic-induced 29 perturbation of the gut ecosystem is associated with transition to toxin-mediated disease. -

Collateral Glucose-Utlizing Pathwaya in Diabetic Polyneuropathy
International Journal of Molecular Sciences Review Collateral Glucose-Utlizing Pathwaya in Diabetic Polyneuropathy Hiroki Mizukami 1,* and Sho Osonoi 1,2 1 Department Pathology and Molecular Medicine, Hirosaki University Graduate School of Medicine, 5 Zaifu-cho, Hirosaki, Aomori 036-8562, Japan; [email protected] 2 Department of Endocrinology and Metabolism, Hirosaki University Graduate School of Medicine, 5 Zaifu-cho, Hirosaki, Aomori 036-8562, Japan * Correspondence: [email protected]; Tel.: +81-172-39-5025 Abstract: Diabetic polyneuropathy (DPN) is the most common neuropathy manifested in diabetes. Symptoms include allodynia, pain, paralysis, and ulcer formation. There is currently no established radical treatment, although new mechanisms of DPN are being vigorously explored. A pathophysio- logical feature of DPN is abnormal glucose metabolism induced by chronic hyperglycemia in the peripheral nerves. Particularly, activation of collateral glucose-utilizing pathways such as the polyol pathway, protein kinase C, advanced glycation end-product formation, hexosamine biosynthetic pathway, pentose phosphate pathway, and anaerobic glycolytic pathway are reported to contribute to the onset and progression of DPN. Inhibitors of aldose reductase, a rate-limiting enzyme involved in the polyol pathway, are the only compounds clinically permitted for DPN treatment in Japan, although their efficacies are limited. This may indicate that multiple pathways can contribute to the pathophysiology of DPN. Comprehensive metabolic analysis may help to elucidate global changes in the collateral glucose-utilizing pathways during the development of DPN, and highlight therapeutic targets in these pathways. Keywords: diabetic polyneuropathy; glycolysis; oxidative stress; polyol pathway; hexosamine biosynthetic pathway; advanced glycation end-products; PKC; glucosamine; pentose phosphate pathway; anaerobic glycolytic pathway Citation: Mizukami, H.; Osonoi, S. -

Development of Potent and Selective Inhibitors of Aldo−Keto Reductase
Article pubs.acs.org/jmc Development of Potent and Selective Inhibitors of Aldo−Keto Reductase 1C3 (Type 5 17β-Hydroxysteroid Dehydrogenase) Based on N-Phenyl-Aminobenzoates and Their Structure−Activity Relationships † § ‡ § † † † ‡ Adegoke O. Adeniji, , Barry M. Twenter, , Michael C. Byrns, Yi Jin, Mo Chen, Jeffrey D. Winkler,*, † and Trevor M. Penning*, † Department of Pharmacology and Center of Excellence in Environmental Toxicology, Perelman School of Medicine, University of Pennsylvania, 130C John Morgan Building, 3620 Hamilton Walk, Philadelphia, Pennsylvania 19104-6084, United States ‡ Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104, United States *S Supporting Information ABSTRACT: Aldo−keto reductase 1C3 (AKR1C3; type 5 17β-hydroxy- steroid dehydrogenase) is overexpressed in castration resistant prostate cancer (CRPC) and is implicated in the intratumoral biosynthesis of testosterone and 5α-dihydrotestosterone. Selective AKR1C3 inhibitors are required because compounds should not inhibit the highly related AKR1C1 and AKR1C2 isoforms which are involved in the inactivation of 5α-dihydrotestosterone. NSAIDs, N-phenylanthranilates in particular, are potent but nonselective AKR1C3 inhibitors. Using flufenamic acid, 2-{[3-(trifluoromethyl)phenyl]amino}- benzoic acid, as lead compound, five classes of structural analogues were synthesized and evaluated for AKR1C3 inhibitory potency and selectivity. Structure−activity relationship (SAR) studies revealed that a meta-carboxylic acid group relative to the amine conferred pronounced AKR1C3 selectivity without loss of potency, while electron withdrawing groups on the phenylamino B-ring were optimal for AKR1C3 inhibition. Lead compounds did not inhibit COX-1 or COX-2 but blocked the AKR1C3 mediated production of testosterone in LNCaP-AKR1C3 cells. These compounds offer promising leads toward new therapeutics for CRPC. -

Bioinformatic and Modelling Approaches for a System-Level Understanding of Oxidative Stress Toxicity Elias Zgheib
Bioinformatic and modelling approaches for a system-level understanding of oxidative stress toxicity Elias Zgheib To cite this version: Elias Zgheib. Bioinformatic and modelling approaches for a system-level understanding of oxidative stress toxicity. Quantitative Methods [q-bio.QM]. Université de Technologie de Compiègne, 2018. English. NNT : 2018COMP2464. tel-02088169 HAL Id: tel-02088169 https://tel.archives-ouvertes.fr/tel-02088169 Submitted on 2 Apr 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Par Elias ZGHEIB Bioinformatic and modelling approaches for a system- level understanding of oxidative stress toxicity Thèse présentée pour l’obtention du grade de Docteur de l’UTC Soutenue le 18 décembre 2018 Spécialité : Bio-ingénierie et Mathématiques Appliquées : Unité de Recherche Biomécanique et Bio-ingénierie (UMR-7338) D2464 BIOINFORMATIC AND MODELLING APPROACHES FOR A SYSTEM-LEVEL UNDERSTANDING OF OXIDATIVE STRESS TOXICITY A THESIS SUBMITTED TO THE UNIVERSITE DE TECHNOLOGIE DE COMPIEGNE SORBONNE UNIVERSITES LABORATOIRE DE BIO-MECANIQUE ET BIOINGENIERIE UMR CNRS 7338 – BMBI 18TH OF DECEMBER 2018 For the degree of Doctor Spécialité : Bio-ingénierie et Mathématiques Appliquées Elias ZGHEIB SUPERVISED BY Prof. Frédéric Y. BOIS JURY MEMBERS Mme. Karine AUDOUZE Rapporteur Mr. -
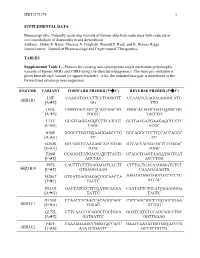
Data Supplement
JPET#173179 1 SUPPLEMENTAL DATA Manuscript title: Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin Authors: Onkar S. Bains, Thomas A. Grigliatti, Ronald E. Reid, and K. Wayne Riggs Journal name: Journal of Pharmacology and Experimental Therapeutics TABLES Supplemental Table 1—Primers for creating non-synonymous single nucleotide polymorphic variants of human AKRs and CBR4 using site-directed mutagenesis. The base pair mutation is given beneath each variant [in square brackets]. Also, the mutated base pair is underlined in the forward and reverse primer sequences. ENZYME VARIANT FORWARD PRIMER (5'3') REVERSE PRIMER (5'3') I15F CAAGATGCCCTTCCTGGGGTT CCAACCCCAGGAAGGGCATC AKR1B1 [AT] GG TTG H42L CGGGTACCGCCTCATCGACTG GGGCACAGTCGATGAGGCGG [AT] TGCCC TACCCG L73V GCGTGAGGAGGTCTTCATCGT GCTGACGATGAAGACCTCCTC [CG] CAGC ACGC K90E GGGCCTGGTGGAAGGAGCCTG GGCAGGCTCCTTCCACCAGGC [AG] CC CC G204S GCCAGTCCAAAAGCATCGTGG GTCACCACGATGCTTTTGGAC [GA] TGAC TGGC T288I CCAGGATATGACCATCTTACTC GTAGCTGAGTAAGATGGTCAT [CT] AGCTAC ATCCTGG P87S CACTTTCTTTGAGAGATCCCTT CTTTCCTCACAAGGGATCTCT AKR1B10 [CT] GTGAGGAAAG CAAAGAAAGTG M286T GTGATGAGGAGACGGCAACCA GAGTATGGTTGCCGTCTCCTC [TC] TACTC ATCAC N313D GACTATCCCTTCGATGCAGAA CAATATTCTGCATCGAAGGGA [AG] TATTG TAGTC R170H CCAACTTCAACCACAGGCAGC CTCCAGCTGCCTGTGGTTGAA AKR1C1 [GA] TGGAG GTTGG Q172L CTTCAACCGCAGGCTGCTGGA GGATCATCTCCAGCAGCCTGC [AT] GATGATCC GGTTGAAG F46Y CAATAGAAGCCGGGTACCACC GAATCAATATGGTGGTACCCG AKR1C2 [TA] ATATTTGATTC GCTTCTATTG JPET#173179