Werner Syndrome Helicase Is a Selective Vulnerability Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Structure and Function of the Human Recq DNA Helicases
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2005 Structure and function of the human RecQ DNA helicases Garcia, P L Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-34420 Dissertation Published Version Originally published at: Garcia, P L. Structure and function of the human RecQ DNA helicases. 2005, University of Zurich, Faculty of Science. Structure and Function of the Human RecQ DNA Helicases Dissertation zur Erlangung der naturwissenschaftlichen Doktorw¨urde (Dr. sc. nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultat¨ der Universitat¨ Z ¨urich von Patrick L. Garcia aus Unterseen BE Promotionskomitee Prof. Dr. Josef Jiricny (Vorsitz) Prof. Dr. Ulrich H ¨ubscher Dr. Pavel Janscak (Leitung der Dissertation) Z ¨urich, 2005 For my parents ii Summary The RecQ DNA helicases are highly conserved from bacteria to man and are required for the maintenance of genomic stability. All unicellular organisms contain a single RecQ helicase, whereas the number of RecQ homologues in higher organisms can vary. Mu- tations in the genes encoding three of the five human members of the RecQ family give rise to autosomal recessive disorders called Bloom syndrome, Werner syndrome and Rothmund-Thomson syndrome. These diseases manifest commonly with genomic in- stability and a high predisposition to cancer. However, the genetic alterations vary as well as the types of tumours in these syndromes. Furthermore, distinct clinical features are observed, like short stature and immunodeficiency in Bloom syndrome patients or premature ageing in Werner Syndrome patients. Also, the biochemical features of the human RecQ-like DNA helicases are diverse, pointing to different roles in the mainte- nance of genomic stability. -

Paul Modrich Howard Hughes Medical Institute and Department of Biochemistry, Duke University Medical Center, Durham, North Carolina, USA
Mechanisms in E. coli and Human Mismatch Repair Nobel Lecture, December 8, 2015 by Paul Modrich Howard Hughes Medical Institute and Department of Biochemistry, Duke University Medical Center, Durham, North Carolina, USA. he idea that mismatched base pairs occur in cells and that such lesions trig- T ger their own repair was suggested 50 years ago by Robin Holliday in the context of genetic recombination [1]. Breakage and rejoining of DNA helices was known to occur during this process [2], with precision of rejoining attributed to formation of a heteroduplex joint, a region of helix where the two strands are derived from the diferent recombining partners. Holliday pointed out that if this heteroduplex region should span a genetic diference between the two DNAs, then it will contain one or more mismatched base pairs. He invoked processing of such mismatches to explain the recombination-associated phenomenon of gene conversion [1], noting that “If there are enzymes which can repair points of damage in DNA, it would seem possible that the same enzymes could recognize the abnormality of base pairing, and by exchange reactions rectify this.” Direct evidence that mismatches provoke a repair reaction was provided by bacterial transformation experiments [3–5], and our interest in this efect was prompted by the Escherichia coli (E. coli) work done in Matt Meselson’s lab at Harvard. Using artifcially constructed heteroduplex DNAs containing multiple mismatched base pairs, Wagner and Meselson [6] demonstrated that mismatches elicit a repair reaction upon introduction into the E. coli cell. Tey also showed that closely spaced mismatches, mismatches separated by a 1000 base pairs or so, are usually repaired on the same DNA strand. -

A Dissertation Entitled the Role of Base Excision Repair And
A Dissertation Entitled The Role of Base Excision Repair and Mismatch Repair Proteins in the Processing of Cisplatin Interstrand Cross-Links. by Akshada Sawant Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biomedical Science Dr. Stephan M. Patrick, Committee Chair Dr. Kandace Williams, Committee Member Dr. William Maltese, Committee Member Dr. Manohar Ratnam, Committee Member Dr. David Giovannucci, Committee Member Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo August 2014 Copyright 2014, Akshada Sawant This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of The Role of Base Excision Repair and Mismatch Repair Proteins in the Processing of Cisplatin Interstrand Cross-Links By Akshada Sawant Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biomedical Science The University of Toledo August 2014 Cisplatin is a well-known anticancer agent that forms a part of many combination chemotherapeutic treatments used against a variety of human cancers. Despite successful treatment, the development of resistance is the major limitation of the cisplatin based therapy. Base excision repair modulates cisplatin cytotoxicity. Moreover, mismatch repair deficiency gives rise to cisplatin resistance and leads to poor prognosis of the disease. Various models have been proposed to explain this low level of resistance caused due to loss of MMR proteins. In our previous studies, we have shown that BER processing of the cisplatin ICLs is mutagenic. Our studies showed that these mismatches lead to the activation and the recruitment of mismatch repair proteins. -

DNA Repair with Its Consequences (E.G
Cell Science at a Glance 515 DNA repair with its consequences (e.g. tolerance and pathways each require a number of apoptosis) as well as direct correction of proteins. By contrast, O-alkylated bases, Oliver Fleck* and Olaf Nielsen* the damage by DNA repair mechanisms, such as O6-methylguanine can be Department of Genetics, Institute of Molecular which may require activation of repaired by the action of a single protein, Biology, University of Copenhagen, Øster checkpoint pathways. There are various O6-methylguanine-DNA Farimagsgade 2A, DK-1353 Copenhagen K, Denmark forms of DNA damage, such as base methyltransferase (MGMT). MGMT *Authors for correspondence (e-mail: modifications, strand breaks, crosslinks removes the alkyl group in a suicide fl[email protected]; [email protected]) and mismatches. There are also reaction by transfer to one of its cysteine numerous DNA repair pathways. Each residues. Photolyases are able to split Journal of Cell Science 117, 515-517 repair pathway is directed to specific Published by The Company of Biologists 2004 covalent bonds of pyrimidine dimers doi:10.1242/jcs.00952 types of damage, and a given type of produced by UV radiation. They bind to damage can be targeted by several a UV lesion in a light-independent Organisms are permanently exposed to pathways. Major DNA repair pathways process, but require light (350-450 nm) endogenous and exogenous agents that are mismatch repair (MMR), nucleotide as an energy source for repair. Another damage DNA. If not repaired, such excision repair (NER), base excision NER-independent pathway that can damage can result in mutations, diseases repair (BER), homologous recombi- remove UV-induced damage, UVER, is and cell death. -

Complex Interacts with WRN and Is Crucial to Regulate Its Response to Replication Fork Stalling
Oncogene (2012) 31, 2809–2823 & 2012 Macmillan Publishers Limited All rights reserved 0950-9232/12 www.nature.com/onc ORIGINAL ARTICLE The RAD9–RAD1–HUS1 (9.1.1) complex interacts with WRN and is crucial to regulate its response to replication fork stalling P Pichierri, S Nicolai, L Cignolo1, M Bignami and A Franchitto Department of Environment and Primary Prevention, Istituto Superiore di Sanita`, Rome, Italy The WRN protein belongs to the RecQ family of DNA preference toward substrates that mimic structures helicases and is implicated in replication fork restart, but associated with stalled replication forks (Brosh et al., how its function is regulated remains unknown. We show 2002; Machwe et al., 2006) and WS cells exhibit that WRN interacts with the 9.1.1 complex, one of enhanced instability at common fragile sites, chromo- the central factors of the replication checkpoint. This somal regions especially prone to replication fork interaction is mediated by the binding of the RAD1 stalling (Pirzio et al., 2008). How WRN favors recovery subunit to the N-terminal region of WRN and is of stalled forks and prevents DNA breakage upon instrumental for WRN relocalization in nuclear foci and replication perturbation is not fully understood. It has its phosphorylation in response to replication arrest. We been suggested that WRN might facilitate replication also find that ATR-dependent WRN phosphorylation restart by either promoting recombination or processing depends on TopBP1, which is recruited by the 9.1.1 intermediates at stalled forks in a way that counteracts complex in response to replication arrest. Finally, we unscheduled recombination (Franchitto and Pichierri, provide evidence for a cooperation between WRN and 2004; Pichierri, 2007; Sidorova, 2008). -

Genome Instability in Secondary Solid Tumors Developing After Radiotherapy of Bilateral Retinoblastoma
Oncogene (2001) 20, 8092 ± 8099 ã 2001 Nature Publishing Group All rights reserved 0950 ± 9232/01 $15.00 www.nature.com/onc Genome instability in secondary solid tumors developing after radiotherapy of bilateral retinoblastoma Sandrine-He leÁ ne LefeÁ vre1, Nicolas Vogt1, Anne-Marie Dutrillaux1, Laurent Chauveinc2, Dominique Stoppa-Lyonnet3, FrancËois Doz4, Laurence Desjardins5, Bernard Dutrillaux1,6, Sylvie Chevillard6 and Bernard Malfoy*,1 1Institut Curie ± CNRS UMR 147, 26 rue d'Ulm 75248 Paris Cedex 05, France; 2Institut Curie, Service de RadiotheÂrapie, 26 rue d'Ulm 75248 Paris Cedex 05, France; 3Institut Curie, Service de GeÂneÂtique Oncologique, 26 rue d'Ulm 75248 Paris Cedex 05, France; 4Institut Curie, Service de PeÂdiatrie, 26 rue d'Ulm 75248 Paris Cedex 05, France; 5Institut Curie, Service d'Ophtalmologie, 26 rue d'Ulm 75248 Paris Cedex 05, France; 6CEA, DSV DRR, 60 avenue du GeÂneÂral Leclerc 92265 Fontenay-aux-Roses, France Genome alterations of seven secondary tumors (®ve these cancers and the diculty in collecting a series of osteosarcomas, one malignant peripheral sheath nerve cases in clearly de®ned context. tumor, one leiomyosarcoma) occurring in the ®eld of It is well established that therapeutic irradiation can irradiation of patients treated for bilateral retinoblasto- induce secondary malignancies within or at the margin ma have been studied. These patients were predisposed to of the radiation ®eld after a long latent period. These develop radiation-induced tumors because of the presence tumors have dierent histology from the primary of a germ line mutation in the retinoblastoma gene lesions, with sarcomas being common (Robinson et (RB1). Tumor cells were characterized by a high al., 1988). -

Arsenic Disruption of DNA Damage Responses—Potential Role in Carcinogenesis and Chemotherapy
Biomolecules 2015, 5, 2184-2193; doi:10.3390/biom5042184 OPEN ACCESS biomolecules ISSN 2218-273X www.mdpi.com/journal/biomolecules/ Review Arsenic Disruption of DNA Damage Responses—Potential Role in Carcinogenesis and Chemotherapy Clarisse S. Muenyi 1, Mats Ljungman 2 and J. Christopher States 3,* 1 Department of Pharmacology and Toxicology, University of Louisville School of Medicine, Louisville, KY 40292, USA; E-Mail: [email protected] 2 Departments of Radiation Oncology and Environmental Health Sciences, University of Michigan, Ann Arbor, MI 48109-2800, USA; E-Mail: [email protected] 3 Department of Pharmacology and Toxicology, University of Louisville School of Medicine, Louisville, KY 40292, USA * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-502-852-5347; Fax: +1-502-852-3123. Academic Editors: Wolf-Dietrich Heyer, Thomas Helleday and Fumio Hanaoka Received: 14 August 2015 / Accepted: 9 September 2015 / Published: 24 September 2015 Abstract: Arsenic is a Class I human carcinogen and is widespread in the environment. Chronic arsenic exposure causes cancer in skin, lung and bladder, as well as in other organs. Paradoxically, arsenic also is a potent chemotherapeutic against acute promyelocytic leukemia and can potentiate the cytotoxic effects of DNA damaging chemotherapeutics, such as cisplatin, in vitro. Arsenic has long been implicated in DNA repair inhibition, cell cycle disruption, and ubiquitination dysregulation, all negatively impacting the DNA damage response and potentially contributing to both the carcinogenic and chemotherapeutic potential of arsenic. Recent studies have provided mechanistic insights into how arsenic interferes with these processes including disruption of zinc fingers and suppression of gene expression. -

Curriculum Vitae
CURRICULUM VITAE NAME: Patricia Lynn Opresko BUSINESS ADDRESS: University of Pittsburgh Graduate School of Public Health Department of Environmental and UPMC Hillman Cancer Center 5117 Centre Avenue, Suite 2.6a Pittsburgh, PA15213-1863 Phone: 412-623-7764 Fax: 412-623-7761 E-mail: [email protected] EDUCATION AND TRAINING Undergraduate 1990 - 1994 DeSales University B.S., 1994 Chemistry and Center Valley, PA Biology Graduate 1994 - 2000 Pennsylvania State Ph.D., 2000 Biochemistry and University, College of Molecular Biology Medicine, Hershey, PA Post-Graduate 3/2000 - 5/2000 Pennsylvania State Postdoctoral Dr. Kristin Eckert, University, College of Fellow Mutagenesis and Medicine, Jake Gittlen Cancer etiology Cancer Research Institute Hershey, PA 2000-2005 National Institute on IRTA Postdoctoral Dr. Vilhelm Bohr Aging, National Fellow Molecular Institutes of Health, Gerontology and Baltimore, MD DNA Repair 1 APPOINTMENTS AND POSITIONS Academic 8/1/2018 – Co-leader Genome Stability Program, UPMC present Hillman Cancer Center 5/1/2018- Tenured Professor Pharmacology and Chemical Biology, present School of Medicine, University of Pittsburgh, Pittsburgh, PA 2/1/2018- Tenured Professor Environmental and Occupational Health, present Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA 2014 – Tenured Associate Environmental and Occupational Health, 1/31/2018 Professor Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA 2005 - 2014 Assistant Professor Environmental and Occupational Health, Graduate School -

Genomic Instability Promoted by Overexpression of Mismatch Repair Factors in Yeast: a Model for Understanding Cancer Progression
HIGHLIGHTED ARTICLE | INVESTIGATION Genomic Instability Promoted by Overexpression of Mismatch Repair Factors in Yeast: A Model for Understanding Cancer Progression Ujani Chakraborty,* Timothy A. Dinh,† and Eric Alani*,1 *Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York 14853-2703 and †Curriculum in Genetics and Molecular Biology, Biological and Biomedical Sciences Program, School of Medicine, University of North Carolina, Chapel Hill, North Carolina 27599 ORCID ID: 0000-0002-5011-9339 (E.A.) ABSTRACT Mismatch repair (MMR) proteins act in spellchecker roles to excise misincorporation errors that occur during DNA replication. Curiously, large-scale analyses of a variety of cancers showed that increased expression of MMR proteins often correlated with tumor aggressiveness, metastasis, and early recurrence. To better understand these observations, we used The Cancer Genome Atlas and Gene Expression across Normal and Tumor tissue databases to analyze MMR protein expression in cancers. We found that the MMR genes MSH2 and MSH6 are overexpressed more frequently than MSH3, and that MSH2 and MSH6 are often cooverex- pressed as a result of copy number amplifications of these genes. These observations encouraged us to test the effects of upregulating MMR protein levels in baker’s yeast, where we can sensitively monitor genome instability phenotypes associated with cancer initiation and progression. Msh6 overexpression (two- to fourfold) almost completely disrupted mechanisms that prevent recombination be- tween divergent DNA sequences by interacting with the DNA polymerase processivity clamp PCNA and by sequestering the Sgs1 helicase. Importantly, cooverexpression of Msh2 and Msh6 (eightfold) conferred, in a PCNA interaction-dependent manner, several genome instability phenotypes including increased mutation rate, increased sensitivity to the DNA replication inhibitor HU and the DNA-damaging agents MMS and 4-nitroquinoline N-oxide, and elevated loss-of-heterozygosity. -

Epigenetic Regulation of DNA Repair Genes and Implications for Tumor Therapy ⁎ ⁎ Markus Christmann , Bernd Kaina
Mutation Research-Reviews in Mutation Research xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Mutation Research-Reviews in Mutation Research journal homepage: www.elsevier.com/locate/mutrev Review Epigenetic regulation of DNA repair genes and implications for tumor therapy ⁎ ⁎ Markus Christmann , Bernd Kaina Department of Toxicology, University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany ARTICLE INFO ABSTRACT Keywords: DNA repair represents the first barrier against genotoxic stress causing metabolic changes, inflammation and DNA repair cancer. Besides its role in preventing cancer, DNA repair needs also to be considered during cancer treatment Genotoxic stress with radiation and DNA damaging drugs as it impacts therapy outcome. The DNA repair capacity is mainly Epigenetic silencing governed by the expression level of repair genes. Alterations in the expression of repair genes can occur due to tumor formation mutations in their coding or promoter region, changes in the expression of transcription factors activating or Cancer therapy repressing these genes, and/or epigenetic factors changing histone modifications and CpG promoter methylation MGMT Promoter methylation or demethylation levels. In this review we provide an overview on the epigenetic regulation of DNA repair genes. GADD45 We summarize the mechanisms underlying CpG methylation and demethylation, with de novo methyl- TET transferases and DNA repair involved in gain and loss of CpG methylation, respectively. We discuss the role of p53 components of the DNA damage response, p53, PARP-1 and GADD45a on the regulation of the DNA (cytosine-5)- methyltransferase DNMT1, the key enzyme responsible for gene silencing. We stress the relevance of epigenetic silencing of DNA repair genes for tumor formation and tumor therapy. -

Expression of MSH2 and MSH6 on a Tissue Microarray in Patients with Osteosarcoma
ANTICANCER RESEARCH 34: 6961-6972 (2014) Expression of MSH2 and MSH6 on a Tissue Microarray in Patients with Osteosarcoma THORSTEN JENTZSCH1,2, BERNHARD ROBL1, MAREN HUSMANN1, BEATA BODE-LESNIEWSKA3 and BRUNO FUCHS1 1Laboratory for Orthopedic Research, Department of Orthopedics, Balgrist University Hospital, Zürich, Switzerland; 2Division of Trauma Surgery, Department of Surgery, University Hospital Zürich, Zürich, Switzerland; 3Institute of Surgical Pathology, University Hospital Zürich, Zürich, Switzerland Abstract. Background/Aim: Reliable prognostic factors for and head (4, 5). Following neoadjuvant chemotherapy, surgical the outcome of patients with osteosarcoma (OS) remain elusive. tumor resections provide tissue specimens for the assessment We analyzed the relationship between immunohistochemical of tumor necrosis rate, histology, grading and evaluation of expression of deoxyribonucleic acid (DNA) mismatch repair tumor biomarkers before further adjuvant chemotherapy may (MMR) proteins, MutS protein homolog 2 (MSH2) and MSH6 be initiated (6-8). Despite advances in therapeutic approaches, using a tissue microarray (TMA) with respect to OS patient five-year overall survival rates have been reported as 78% (9) demographics and survival time. Materials and Methods: We and are usually lower for non-responders to chemotherapy retrieved tumor tissue specimens from bone tissue originating (10) and patients with metastasis (11, 12). from surgical primary tumor specimens of OS patients to Albeit the limited number of studies (8, 13-17) on tumor generate a TMA and stained sections with antibodies against biomarkers, these are important for treatment decisions, MSH2 and MSH6. Results: Tumor resections of 67 patients patient discrimination regarding the response to with a mean follow-up of 98 months were evaluated. MSH2 chemotherapy and the incidence of metastasis, prediction of was expressed in nine (13%), MSH6 in ten (15%) and patient survival times and patient education (18-21). -
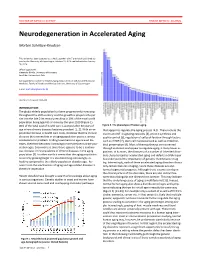
Neurodegeneration in Accelerated Aging
DOCTOR OF MEDICAL SCIENCE DANISH MEDICAL JOURNAL Neurodegeneration in Accelerated Aging Morten Scheibye-Knudsen This review has been accepted as a thesis together with 7 previously published pa- pers by the University of Copenhagen, October 16, 2014 and defended on January 14, 2016 Official opponents: Alexander Bürkle, University of Konstanz Lars Eide, University of Oslo Correspondence: Center for Healthy Aging, Department of Cellular and Molecular Medicine, Faculty of Health and Medical Sciences, University of Copenhagen E-mail: [email protected] Dan Med J 2016;63(11):B5308 INTRODUCTION The global elderly population has been progressively increasing throughout the 20th century and this growth is projected to per- sist into the late 21st century resulting in 20% of the total world population being aged 65 or more by the year 2100 (Figure 1). 80% of the total cost of health care is accrued after 40 years of Figure 2. The phenotype of human aging. age where chronic diseases become prevalent [1, 2]. With an ex- that appear to regulate the aging process [4,5]. These include the ponential increase in health care costs, it follows that the chronic insulin and IGF-1 signaling cascades [4], protein synthesis and diseases that accumulate in an aging population poses a serious quality control [6], regulation of cell proliferation through factors socioeconomic problem. Finding treatments to age related dis- such as mTOR [7], stem cell maintenance 8 as well as mitochon- eases, therefore becomes increasingly more pertinent as the pop- drial preservation [9]. Most of these pathways are conserved ulation ages. Even more so since there appears to be a continu- through evolution and appear to regulate aging in many lower or- ous increase in the prevalence of chronic diseases in the aging ganisms.