Chromosome Homology Between Mouse and Three Muridae Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

13914444D46c0aa91d02e31218
2 Breeding of wild and some domestic animals at regional zoological institutions in 2013 3 РЫБЫ P I S C E S ВОББЕЛОНГООБРАЗНЫЕ ORECTOLOBIFORMES Сем. Азиатские кошачьи акулы (Бамбуковые акулы) – Hemiscyllidae Коричневополосая бамбуковая акула – Chiloscyllium punctatum Brownbanded bambooshark IUCN (NT) Sevastopol 20 ХВОСТОКОЛООБРАЗНЫЕ DASYATIFORMES Сем. Речные хвостоколы – Potamotrygonidae Глазчатый хвостокол (Моторо) – Potamotrygon motoro IUCN (DD) Ocellate river stingray Sevastopol - ? КАРПООБРАЗНЫЕ CYPRINIFORMES Сем. Цитариновые – Citharinidae Серебристый дистиход – Distichodusaffinis (noboli) Silver distichodus Novosibirsk 40 Сем. Пираньевые – Serrasalmidae Серебристый метиннис – Metynnis argenteus Silver dollar Yaroslavl 10 Обыкновенный метиннис – Metynnis schreitmuelleri (hypsauchen) Plainsilver dollar Nikolaev 4; Novosibirsk 100; Kharkov 20 Пятнистый метиннис – Metynnis maculatus Spotted metynnis Novosibirsk 50 Пиранья Наттерера – Serrasalmus nattereri Red piranha Novosibirsk 80; Kharkov 30 4 Сем. Харацидовые – Characidae Красноплавничный афиохаракс – Aphyocharax anisitsi (rubripinnis) Bloodfin tetra Киев 5; Perm 10 Парагвайский афиохаракс – Aphyocharax paraquayensis Whitespot tetra Perm 11 Рубиновый афиохаракс Рэтбина – Aphyocharax rathbuni Redflank bloodfin Perm 10 Эквадорская тетра – Astyanax sp. Tetra Perm 17 Слепая рыбка – Astyanax fasciatus mexicanus (Anoptichthys jordani) Mexican tetra Kharkov 10 Рублик-монетка – Ctenobrycon spilurus (+ С. spilurusvar. albino) Silver tetra Kharkov 20 Тернеция (Траурная тетра) – Gymnocorymbus -

Sexual Dimorphism in Brain Transcriptomes of Amami Spiny Rats (Tokudaia Osimensis): a Rodent Species Where Males Lack the Y Chromosome Madison T
Ortega et al. BMC Genomics (2019) 20:87 https://doi.org/10.1186/s12864-019-5426-6 RESEARCHARTICLE Open Access Sexual dimorphism in brain transcriptomes of Amami spiny rats (Tokudaia osimensis): a rodent species where males lack the Y chromosome Madison T. Ortega1,2, Nathan J. Bivens3, Takamichi Jogahara4, Asato Kuroiwa5, Scott A. Givan1,6,7,8 and Cheryl S. Rosenfeld1,2,8,9* Abstract Background: Brain sexual differentiation is sculpted by precise coordination of steroid hormones during development. Programming of several brain regions in males depends upon aromatase conversion of testosterone to estrogen. However, it is not clear the direct contribution that Y chromosome associated genes, especially sex- determining region Y (Sry), might exert on brain sexual differentiation in therian mammals. Two species of spiny rats: Amami spiny rat (Tokudaia osimensis) and Tokunoshima spiny rat (T. tokunoshimensis) lack a Y chromosome/Sry, and these individuals possess an XO chromosome system in both sexes. Both Tokudaia species are highly endangered. To assess the neural transcriptome profile in male and female Amami spiny rats, RNA was isolated from brain samples of adult male and female spiny rats that had died accidentally and used for RNAseq analyses. Results: RNAseq analyses confirmed that several genes and individual transcripts were differentially expressed between males and females. In males, seminal vesicle secretory protein 5 (Svs5) and cytochrome P450 1B1 (Cyp1b1) genes were significantly elevated compared to females, whereas serine (or cysteine) peptidase inhibitor, clade A, member 3 N (Serpina3n) was upregulated in females. Many individual transcripts elevated in males included those encoding for zinc finger proteins, e.g. -

Cricetidae, Sigmodontinae): Searching for Ancestral Phylogenetic Traits
RESEARCH ARTICLE Extensive Chromosomal Reorganization in the Evolution of New World Muroid Rodents (Cricetidae, Sigmodontinae): Searching for Ancestral Phylogenetic Traits Adenilson Leão Pereira1, Stella Miranda Malcher1, Cleusa Yoshiko Nagamachi1,2, Patricia Caroline Mary O’Brien3, Malcolm Andrew Ferguson-Smith3, Ana Cristina Mendes- Oliveira4, Julio Cesar Pieczarka1,2* 1 Laboratório de Citogenética, Centro de Estudos Avançados da Biodiversidade, ICB, Universidade Federal do Pará, Belém, Pará, Brasil, 2 CNPq Researcher, Brasília, Brasil, 3 Cambridge Resource Center for Comparative Genomics, Department of Veterinary Medicine, University of Cambridge, Cambridge, United Kingdom, 4 Laboratório de Zoologia e Ecologia de Vertebrados, ICB, Universidade Federal do Pará, Belém, Pará, Brasil * [email protected] OPEN ACCESS Citation: Pereira AL, Malcher SM, Nagamachi CY, O’Brien PCM, Ferguson-Smith MA, Mendes-Oliveira Abstract AC, et al. (2016) Extensive Chromosomal Reorganization in the Evolution of New World Muroid Sigmodontinae rodents show great diversity and complexity in morphology and ecology. Rodents (Cricetidae, Sigmodontinae): Searching for This diversity is accompanied by extensive chromosome variation challenging attempts to Ancestral Phylogenetic Traits. PLoS ONE 11(1): reconstruct their ancestral genome. The species Hylaeamys megacephalus–HME (Oryzo- e0146179. doi:10.1371/journal.pone.0146179 myini, 2n = 54), Necromys lasiurus—NLA (Akodontini, 2n = 34) and Akodon sp.–ASP (Ako- Editor: Riccardo Castiglia, Universita degli Studi di dontini, 2n = 10) have extreme diploid numbers that make it difficult to understand the Roma La Sapienza, ITALY rearrangements that are responsible for such differences. In this study we analyzed these Received: June 5, 2015 changes using whole chromosome probes of HME in cross-species painting of NLA and Accepted: December 13, 2015 ASP to construct chromosome homology maps that reveal the rearrangements between Published: January 22, 2016 species. -

Èíòåãðàöèîííûå Ïðîåêòû Ñî Ðàí Âûï. 7 Sb Ras Integrate
Перейти на страницу с полной версией» Âûï. 7 SB RAS INTEGRATE PROJECTS INTEGRATE RAS SB ÈÍÒÅÃÐÀÖÈÎÍÍÛÅ ÏÐÎÅÊÒÛ ÑÎ ÐÀÍ ÑÎ ÏÐÎÅÊÒÛ ÈÍÒÅÃÐÀÖÈÎÍÍÛÅ Issue 7 Перейти на страницу с полной версией» Перейти на страницу с полной версией» RUSSIAN ÀCÀDEMY O SCIENCES SIBERIAN BRANCH BIODIVERSITY and Dinamic of Ecosystems: Computationl Approaches and Modelling Executive editors: Academician of RAS V.K. Shumny, Yu.I. Shokin Corresponding member of RAS N.A. Kolchanov, A.M. edotov NOVOSIBIRSK PUBLISHING HAUSE O THE SIBERIAN BRANCH O THE RUSSIAN ACADEMY O SCIENCES 2006 Перейти на страницу с полной версией» Перейти на страницу с полной версией» ÐÎÑÑÈÉÑÊÀß ÀÊÀÄÅÌÈß ÍÀÓÊ ÑÈÁÈÐÑÊÎÅ ÎÒÄÅËÅÍÈÅ ÈÍÑÒÈÒÓÒ ÖÈÒÎËÎÃÈÈ È ÃÅÍÅÒÈÊÈ ÈÍÑÒÈÒÓÒ ÂÛ×ÈÑËÈÒÅËÜÍÛÕ ÒÅÕÍÎËÎÃÈÉ ÈÍÑÒÈÒÓÒ ËÅÑÀ èì. Â.Í. ÑÓÊÀ×ÅÂÀ ÈÍÑÒÈÒÓÒ ÑÈÑÒÅÌÀÒÈÊÈ È ÝÊÎËÎÃÈÈ ÆÈÂÎÒÍÛÕ ÖÅÍÒÐÀËÜÍÛÉ ÑÈÁÈÐÑÊÈÉ ÁÎÒÀÍÈ×ÅÑÊÈÉ ÑÀÄ ÍÎÂÎÑÈÁÈÐÑÊÈÉ ÃÎÑÓÄÀÐÑÒÂÅÍÍÛÉ ÓÍÈÂÅÐÑÈÒÅÒ ÁÈÎÐÀÇÍÎÎÁÐÀÇÈÅ È ÄÈÍÀÌÈÊÀ ÝÊÎÑÈÑÒÅÌ: ÈÍÔÎÐÌÀÖÈÎÍÍÛÅ ÒÅÕÍÎËÎÃÈÈ È ÌÎÄÅËÈÐÎÂÀÍÈÅ Îòâåòñòâåííûå ðåäàêòîðû: àêàäåìèêè ÐÀÍ Â.Ê. Øóìíûé, Þ.È. Øîêèí ÷ëåíû-êîððåñïîíäåíòû ÐÀÍ Í.À. Êîë÷àíîâ, À.Ì. Ôåäîòîâ ÍÎÂÎÑÈÁÈÐÑÊ ÈÇÄÀÒÅËÜÑÒÂÎ ÑÈÁÈÐÑÊÎÃÎ ÎÒÄÅËÅÍÈß ÐÎÑÑÈÉÑÊÎÉ ÀÊÀÄÅÌÈÈ ÍÀÓÊ 2006 Перейти на страницу с полной версией» Перейти на страницу с полной версией» ÓÄÊ 574:004.9 ÁÁÊ 28.0+32.97 Á63 Ðåäàêöèîííàÿ êîëëåãèÿ ñåðèè: àêàäåìèê Â.Ì. Òèòîâ (ãëàâíûé ðåäàêòîð), àêàäåìèê Þ.È. Øîêèí, ÷ëåí-êîððåñïîíäåíò ÐÀÍ Â.È. Åâñèêîâ, ÷ëåí-êîððåñïîíäåíò ÐÀÍ Â.Í. Îïàðèí, ÷ëåí-êîððåñïîíäåíò ÐÀÍ Â.À. Ëàìèí, êàíä. ãåîë.-ìèí. íàóê Â.Ä. Åðìèêîâ (îòâåòñòâåííûé ñåêðåòàðü) Ñåðèÿ îñíîâàíà â 2003 ã. Áèîðàçíîîáðàçèå è äèíàìèêà ýêîñèñòåì: èíôîðìàöèîííûå òåõíîëî- Á63 ãèè è ìîäåëèðîâàíèå / îòâ. ðåä. Â.Ê. Øóìíûé, Þ.È. Øîêèí, Í.À. Êîë- ÷àíîâ, À.Ì. Ôåäîòîâ; Ðîñ. -

Sex Determination and Genetic Management in Nile Tilapia Using Genomic Techniques
Sex Determination and Genetic Management in Nile Tilapia using Genomic Techniques A Thesis Submitted for the Degree of Doctor of Philosophy By Taslima Khanam Institute of Aquaculture, Faculty of Natural Sciences, University of Stirling, Stirling, UK January 2017 Acknowledgements My PhD has been an amazing experience and it would have never been achievable without the generous support and guidance that I received from very important and special persons. The first person I would like to express my heartfelt gratitude to my Principal supervisor Prof. Brendan J. McAndrew for accepting me as a PhD student under his scholastic supervision. I would like to say a very big thank you for all the supports and encouragements during the study period. Thank you very much for being there always and quickly sorted when I needed any support, which made my PhD a wonderful experience. I sincerely appreciate your active participation during thesis/manuscript write up and for valuable suggestions. I would like to express my indebtness to my supervisor Dr. David J. Penman wholeheartedly; you have been a tremendous mentor for me. Your kind support, intellectual stimulation, constant valued guidance, judicious constructive criticism and encouragement made my PhD a pleasing experience and enabled me to grow as a researcher. Thank you very much Dave for all the time invested in me to improve my research thinking and keeping your door always open to talk to you, and active participation while I was doing experiments and writing up articles/thesis. Your willingness to introduce me with the renowned scientists has been priceless. I am very pleased that I did my PhD research under both of your supervisions. -
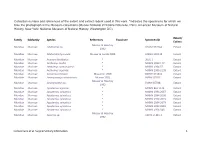
Collection Number and References of the Extant and Extinct Rodent Used in This Work
Collection number and references of the extant and extinct rodent used in this work. *indicates the specimens for which we take the photograph in the Museum collections (Musée National d’Histoire Naturelle, Paris; American Museum of Natural History, New York; National Museum of Natural History, Washington DC). Extant/ Family Subfamily Species References Fossil site Specimen ID Extinct Musser & Headley Muridae Murinae Abditomys sp. USNM 357244 Extant 1992 Muridae Murinae Abelomelomys sevia Musser & Lunde 2009 AMNH 192119 Extant Muridae Murinae Acomys dimidiatus * 2001 3 Extant Muridae Murinae Aethomys hindei * MNHN 1999 177 Extant Muridae Murinae Aethomys namanquesis * MNHN 1964 57 Extant Muridae Murinae Aethomys nigeriae * MNHN 1996 2239 Extant Muridae Murinae Anisomys imitator Missonne 1969 BMNH 471310 Extant Muridae Murinae Anonymomys mindorensis Musser 1981 FMNH 87597 Extant Musser & Headley Muridae Murinae Anonymomys sp. FMNH 87598 Extant 1992 Muridae Murinae Apodemus agrarius * MNHN BL6 1134 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2667 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2668 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2671 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2679 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2681 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 945 Extant Musser & Headley Muridae Murinae Apomys sp. 12971 CLM1-3 Extant 1992 Gomez Cano et al. Supplementary Information 1 Collection number and references of the extant and extinct rodent used in this work. *indicates the specimens for which we take the photograph in the Museum collections (Musée National d’Histoire Naturelle, Paris; American Museum of Natural History, New York; National Museum of Natural History, Washington DC). Extant/ Family Subfamily Species References Fossil site Specimen ID Extinct Musser & Headley Muridae Murinae Archboldomys sp. -

Rediscovery After Thirty Years Since the Last Capture of the Critically Endangered Okinawa Spiny Rat Tokudaia Muenninki Title in the Northern Part of Okinawa Island
Rediscovery after thirty years since the last capture of the critically endangered Okinawa spiny rat Tokudaia muenninki Title in the northern part of Okinawa Island Yamada, Fumio; Kawauchi, Norihiro; Nakata, Katsushi; Abe, Shintaro; Kotaka, Nobuhiko; Takashima, Atsushi; Author(s) Murata, Chie; Kuroiwa, Asato Mammal Study, 35(4), 243-255 Citation https://doi.org/10.3106/041.035.0404 Issue Date 2010-12 Doc URL http://hdl.handle.net/2115/47531 Type article File Information MS35-4_243-255.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP Mammal Study 35: 243–255 (2010) © the Mammalogical Society of Japan Rediscovery after thirty years since the last capture of the critically endangered Okinawa spiny rat Tokudaia muenninki in the northern part of Okinawa Island Fumio Yamada1,*, Norihiro Kawauchi2,**, Katsushi Nakata3, Shintaro Abe4, Nobuhiko Kotaka5, Atsushi Takashima6, Chie Murata7 and Asato Kuroiwa7,8 1 Forestry and Forest Products Research Institute, Tsukuba, Ibaraki 305-8687, Japan 2 Island Ecology Institute, Ginowan, Okinawa 901-2206, Japan 3 Yambaru Wildlife Conservation Center, Ministry of the Environment, Kunigami, Okinawa 905-1413, Japan 4 Naha Nature Conservation Office, Ministry of the Environment, Naha, Okinawa 900-0027, Japan 5 Kyushu Research Center, Forestry and Forest Products Research Institute, Kumamoto, Kumamoto 860-0862, Japan 6 Yona Field, Subtropical Field Science Center, Faculty of Agriculture, University of the Ryukyus, Kunigami, Okinawa 905-1427, Japan 7 Graduate School of Life Science, Hokkaido University, Sapporo, Hokkaido 060-0810, Japan 8 Laboratory of Animal Cytogenetics, Faculty of Science, Hokkaido University, Sapporo, Hokkaido 060-0810, Japan Abstract. The Okinawa spiny rat, Tokudaia muenninki, is a critically endangered species endemic to the northern part of Okinawa Island and may be extinct in the wild as there have been no recent sightings of the animal in its natural habitat. -
Molecular Pattern Recognition of A
Hyperconservation of the N-Formyl Peptide Binding Site of M3: Evidence that M3 Is an Old Eutherian Molecule with Conserved Recognition of a Pathogen-Associated This information is current as Molecular Pattern of September 24, 2021. C. Kuyler Doyle, Beckley K. Davis, Richard G. Cook, Robert R. Rich and John R. Rodgers J Immunol 2003; 171:836-844; ; doi: 10.4049/jimmunol.171.2.836 Downloaded from http://www.jimmunol.org/content/171/2/836 References This article cites 63 articles, 25 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/171/2/836.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 24, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2003 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Hyperconservation of the N-Formyl Peptide Binding Site of M3: Evidence that M3 Is an Old Eutherian Molecule with Conserved Recognition of a Pathogen-Associated Molecular Pattern1 C. -

Comparative Chromosome Painting Map Between Two Ryukyu Spiny Rat Species, Tokudaia Osimensis and Tokudaia Title Tokunoshimensis (Muridae, Rodentia)
Comparative chromosome painting map between two Ryukyu spiny rat species, Tokudaia osimensis and Tokudaia Title tokunoshimensis (Muridae, Rodentia) Author(s) Nakamura, Taro; Kuroiwa, Asato; Nishida-Umehara, Chizuko; Matsubara, Kazumi; Yamada, Fumio; Matsuda, Yoichi Chromosome Research, 15(6), 799-806 Citation https://doi.org/10.1007/s10577-007-1163-9 Issue Date 2007-10 Doc URL http://hdl.handle.net/2115/30262 Rights The original publication is available at www.springerlink.com Type article (author version) File Information CR15-6.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP Comparative chromosome painting map between two Ryukyu spiny rat species, Tokudaia osimensis and Tokudaia tokunoshimensis (Muridae, Rodentia) Taro Nakamura1, Asato Kuroiwa1, 2, Chizuko Nishida-Umehara1, 2, Kazumi Matsubara2, Fumio Yamada3 and Yoichi Matsuda1, 2* 1 Laboratory of Animal Cytogenetics, Graduate School of Science, Hokkaido University, North 10 West 8, Kita-ku, Sapporo 060-0810, Japan; 2 Laboratory of Animal Cytogenetics, Creative Research Initiative “Sousei”, Hokkaido University, North 10 West 8, Kita-ku, Sapporo 060-0810, Japan, Tel: +81-11-706-2619; Fax: +81-11-736-6304; E-mail:[email protected]; 3 Kansai Research Center, Forestry and Forest Products Research Institute, Nagaikyutaro 68, Momoyama, Kyoto 612-0855, Japan *Correspondence Key Words: Tokudaia, comparative chromosome painting, chromosome homology, ancestral karyotype, karyotype evolution Abstract Ryukyu spiny rats (genus Tokudaia) are indigenous species that are confined to three islands of the Nansei Shoto archipelago, Amami-Oshima, Tokunoshima and Okinawa-jima, Japan. Tokudaia tokunoshimensis from Tokunoshima Island and Tokudaia osimensis from Amami-Oshima Island are closely related taxonomically, although their karyotypes are quite different: the diploid chromosome numbers and sex chromosome constitution are 2n=45, X0/X0 for T. -

AZE.Qrk 11/14/05 5:14 PM Page 1
AZE.Qrk 11/14/05 5:14 PM Page 1 Mexico/Isla Socorro (MX34) Photo by Mike Parr ALLIANCE FOR Volcano rabbit Romerolagus diazi Azores bullfinch Pyrrhula murina Mexico/Sur del Valle de México (MX55) Portugal/Azores, east of São Miguel (PO1) Photo by Alejandro Velázquez Zero Extinction Photo by Leo J.R. Boon/Cursorius Pinpointing and Preventing Imminent Extinctions • www.zeroextinction.org Whooping crane Grus americana Asia Minor spiny mouse Costa Rica/Osa US/Aransas National Wildlife Refuge (US2), Acomys cilicicus Peninsula (CR2) Canada/Wood Buffalo National Park (CA2) Turkey/Silifke (TU2) Photo by Mike Parr Photo by USFWS he Alliance for Zero Extinction (AZE), a joint initia- Photo by Ahmet Karatas tive of 52 biodiversity conservation organizations, T aims to prevent extinctions by identifying and safe- guarding key sites where species are in imminent danger of disappearing. The goal of the Alliance is to create a front Flat-backed spider tortoise Pyxis planicauda Madagascar/Menabe Forest (MA13) Dusky gopher frog Rana sevosa line of defense against extinction by eliminating threats and Photo by John Behler, Bronx Zoo/WCS US/Glen's Pond in Mont Nimba viviparous toad Okinawa rail US10 Worthen's sparrow Spizella wortheni De Soto National Forest (US5) Nimbaphrynoides occidentalis Gallirallus okinawae Photo by Suzanne L. Collins/The Center Guinea/Mont Nimba (GU1) Mexico/Saltillo savanna (MX47) Torrey pine Pinus torreyana restoring habitat to allow species populations to rebound. Japan/Yambaru (JA8) Photo by Ray Bieber for North American Herpetology -

A New Species of Ryukyu Spiny Rat, Tokudaia (Muridae: Rodentia), from Tokunoshima Island, Kagoshima Prefecture, Japan
Mammal Study 31: 47–57 (2006) © the Mammalogical Society of Japan A new species of Ryukyu spiny rat, Tokudaia (Muridae: Rodentia), from Tokunoshima Island, Kagoshima Prefecture, Japan Hideki Endo1,* and Kimiyuki Tsuchiya2,** 1 Section of Morphology, Primate Research Institute, Kyoto University, Inuyama, Aichi 484-8506, Japan 2 Laboratory of Wild Animals, Department of Animal Sciences, Faculty of Agriculture, Tokyo University of Agriculture, Atsugi, Kanagawa 243-0034, Japan Abstract. A new species of Ryukyu spiny rat, Tokudaia tokunoshimensis was described in the specimens originating from Tokunoshima Island in the southernmost region of Japan. The populations of Tokudaia are separately distributed only in Amami-Oshima, Tokunoshima, and Okinawa-jima Islands. We have described the osteological and external morphological characteristics and clarified the osteometrical distinctions among the three populations using the skull and skin collections. All external dimensions of head and body length, tail length, hindfoot length and ear length were larger in the Tokunoshima population than in the other two in mean value. The raw osteometrical data separated the Tokunoshima Island population from the other two in all measurements except for the length of molar row in the Okinawa-jima Island population. The principal component analysis also demonstrated that the skulls from the Tokunoshima Island population were morphologically distinct from those of the other populations. Here, from these findings, we point out that the Tokunoshima population of Tokudaia should be regarded as an independent species from the two other populations. Key words: osteology, Ryukyu spiny rat, Tokudaia muenninki, Tokudaia osimensis, Tokunoshima. The Ryukyu spiny rats (genus Tokudaia) are distributed mal DNA (Tsuchiya et al. -

Muennink's Spiny Rat Katherine Sabak First Named As a Separate
Muennink’s Spiny Rat Katherine Sabak First named as a separate species of rodent in 1946, Tokudaia muenninki , also known as Muennink’s spiny rat or the Okinawa spiny rat, lives in the northern region of Yanbaru Forest on Okinawa Island, Japan. It is part of the family Muridae and the genus Tokudaia which is comprised of this and two other species ( T. osimensis and T. tokunoshimensis ); the members of this genus are the closest living relatives of the European field mouse ( Apodemus ). Each of the three species lives only on one specific island; T. muenninki lives on Okinawa Island, T. osimensis lives on the island of Amami Oshima, and T. tokunoshimensis lives on the island of Tokunoshima. A variety of physical and genetic differences among the species result from this early separation. The other two species within the genus are also endangered; the Okinawa spiny rat, however, has been ranked as critically endangered according to the International Union for Conservation of Nature’s list of endangered animals since 2008. (There are at least 27 endangered species living on or near Okinawa Island.) It is the most vulnerable of the species within Tokudaia to decline in population because of the following factors: severe habitat fragmentation due to forestry programs, competition with black rats for resources, and predation by non-native species such as the small Indian mongoose ( Herpestes auropunctatus ) and feral cats ( Felis catus ). It is estimated that the remaining members of the species live in an area of about one square mile within the forest. (The Okinawa spiny rats live at altitudes over 984 feet above sea level.) The most recent sighting and capture of wild specimens was in March 2008; before this discovery, it was assumed that the species was extinct in the wild, as there had been no evidence of their continued existence for thirty years.