HDAC and MAPK/ERK Inhibitors Cooperate to Reduce
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Recombinant MAP2K2 Protein
Recombinant MAP2K2 protein Catalog No: 81332, 81632 Quantity: 20, 1000 µg Expressed In: Baculovirus Concentration: 0.3 µg/µl Source: Human Buffer Contents: Recombinant MAP2K2 protein is supplied in 25 mM HEPES-NaOH pH 7.5, 300 mM NaCl, 10% glycerol, 0.04% Triton X-100, and 0.5 mM TCEP. Background: MAP2K2 (Mitogen-Activated Protein Kinase Kinase Kinase 2) is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase plays a critical role in mitogen growth factor signal transduction. It phosphorylates and activates MAPK1/ERK2 and MAPK2/ERK3. The activation of this kinase itself is dependent on the Ser/Thr phosphorylation by MAP kinase kinase kinases. Mutations in this gene cause cardiofaciocutaneous syndrome (CFC syndrome), a disease characterized by heart defects, cognitive disability, and distinctive facial features similar to those found in Noonan syndrome. Protein Details: Recombinant MAP2K2 protein was expressed in baculovirus expression system as the full length protein (accession number NP_109587.1) with a N-terminal FLAG Tag. The molecular weight of the protein is 45.7 kDa. Application Notes: This product was manufactured as described in Protein Details. Where possible, Active Motif has developed functional or activity assays for Recombinant MAP2K2 protein gel recombinant proteins. Additional characterization such as enzyme kinetic activity 10% SDS-PAGE with Coomassie blue assays, inhibitor screening or other biological activity assays may not have been staining MW: 45.7 kDa performed for every product. All available data for a given product is shown on the lot- Purity: >90% specific Technical Data Sheet. Storage and Guarantee: Recombinant proteins in solution are temperature sensitive and must be stored at -80°C to prevent degradation. -

Tumour-Agnostic Therapy for Pancreatic Cancer and Biliary Tract Cancer
diagnostics Review Tumour-Agnostic Therapy for Pancreatic Cancer and Biliary Tract Cancer Shunsuke Kato Department of Clinical Oncology, Juntendo University Graduate School of Medicine, 2-1-1, Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; [email protected]; Tel.: +81-3-5802-1543 Abstract: The prognosis of patients with solid tumours has remarkably improved with the develop- ment of molecular-targeted drugs and immune checkpoint inhibitors. However, the improvements in the prognosis of pancreatic cancer and biliary tract cancer is delayed compared to other carcinomas, and the 5-year survival rates of distal-stage disease are approximately 10 and 20%, respectively. How- ever, a comprehensive analysis of tumour cells using The Cancer Genome Atlas (TCGA) project has led to the identification of various driver mutations. Evidently, few mutations exist across organs, and basket trials targeting driver mutations regardless of the primary organ are being actively conducted. Such basket trials not only focus on the gate keeper-type oncogene mutations, such as HER2 and BRAF, but also focus on the caretaker-type tumour suppressor genes, such as BRCA1/2, mismatch repair-related genes, which cause hereditary cancer syndrome. As oncogene panel testing is a vital approach in routine practice, clinicians should devise a strategy for improved understanding of the cancer genome. Here, the gene mutation profiles of pancreatic cancer and biliary tract cancer have been outlined and the current status of tumour-agnostic therapy in these cancers has been reported. Keywords: pancreatic cancer; biliary tract cancer; targeted therapy; solid tumours; driver mutations; agonist therapy Citation: Kato, S. Tumour-Agnostic Therapy for Pancreatic Cancer and 1. -

COVID-19—The Potential Beneficial Therapeutic Effects of Spironolactone During SARS-Cov-2 Infection
pharmaceuticals Review COVID-19—The Potential Beneficial Therapeutic Effects of Spironolactone during SARS-CoV-2 Infection Katarzyna Kotfis 1,* , Kacper Lechowicz 1 , Sylwester Drozd˙ zal˙ 2 , Paulina Nied´zwiedzka-Rystwej 3 , Tomasz K. Wojdacz 4, Ewelina Grywalska 5 , Jowita Biernawska 6, Magda Wi´sniewska 7 and Miłosz Parczewski 8 1 Department of Anesthesiology, Intensive Therapy and Acute Intoxications, Pomeranian Medical University in Szczecin, 70-111 Szczecin, Poland; [email protected] 2 Department of Pharmacokinetics and Monitored Therapy, Pomeranian Medical University, 70-111 Szczecin, Poland; [email protected] 3 Institute of Biology, University of Szczecin, 71-412 Szczecin, Poland; [email protected] 4 Independent Clinical Epigenetics Laboratory, Pomeranian Medical University, 71-252 Szczecin, Poland; [email protected] 5 Department of Clinical Immunology and Immunotherapy, Medical University of Lublin, 20-093 Lublin, Poland; [email protected] 6 Department of Anesthesiology and Intensive Therapy, Pomeranian Medical University in Szczecin, 71-252 Szczecin, Poland; [email protected] 7 Clinical Department of Nephrology, Transplantology and Internal Medicine, Pomeranian Medical University, 70-111 Szczecin, Poland; [email protected] 8 Department of Infectious, Tropical Diseases and Immune Deficiency, Pomeranian Medical University in Szczecin, 71-455 Szczecin, Poland; [email protected] * Correspondence: katarzyna.kotfi[email protected]; Tel.: +48-91-466-11-44 Abstract: In March 2020, coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 was declared Citation: Kotfis, K.; Lechowicz, K.; a global pandemic by the World Health Organization (WHO). The clinical course of the disease is Drozd˙ zal,˙ S.; Nied´zwiedzka-Rystwej, unpredictable but may lead to severe acute respiratory infection (SARI) and pneumonia leading to P.; Wojdacz, T.K.; Grywalska, E.; acute respiratory distress syndrome (ARDS). -

TAK1 Mediates Convergence of Cellular Signals for Death and Survival
Apoptosis (2019) 24:3–20 https://doi.org/10.1007/s10495-018-1490-7 REVIEW TAK1 mediates convergence of cellular signals for death and survival Sabreena Aashaq1 · Asiya Batool1 · Khurshid I. Andrabi1 Published online: 4 October 2018 © Springer Science+Business Media, LLC, part of Springer Nature 2018 Abstract TGF-β activated kinase 1, a MAPK kinase kinase family serine threonine kinase has been implicated in regulating diverse range of cellular processes that include embryonic development, differentiation, autophagy, apoptosis and cell survival. TAK1 along with its binding partners TAB1, TAB2 and TAB3 displays a complex pattern of regulation that includes serious crosstalk with major signaling pathways including the C-Jun N-terminal kinase (JNK), p38 MAPK, and I-kappa B kinase complex (IKK) involved in establishing cellular commitments for death and survival. This review also highlights how TAK1 orchestrates regulation of energy homeostasis via AMPK and its emerging role in influencing mTORC1 pathway to regulate death or survival in tandem. Keywords Apoptosis · Autophagy · Cytokine · Inflammatory · Smad TAK1, a multifunctional kinase regulate a wide array of downstream cellular responses [2, 7, 8]. Various branches of MAP kinase pathways including Transforming growth factor-β is a versatile cytokine, regu- the extracellular signal regulated kinase (Erk) ½ [3, 9], p38 lating a wide variety of intracellular signaling pathways. MAPK [10, 11], c-Jun N-Terminal kinase (JNK) [12, 13], The Smad dependent signaling pathway is conventionally phosphatidylinositol-3-kinase/AKT pathway [14, 15] and acknowledged as the traditional pathway promoted by TGF- Rho-like GTPase [16, 17] signaling pathways are included β1 [1]. However, the Smad dependent signaling pathway among the Smad independent pathways. -

Map2k1 and Map2k2 Genes Contribute to the Normal Development of Syncytiotrophoblasts During Placentation
RESEARCH ARTICLE 1363 Development 136, 1363-1374 (2009) doi:10.1242/dev.031872 Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation Valérie Nadeau*, Stéphanie Guillemette*, Louis-François Bélanger, Olivier Jacob, Sophie Roy and Jean Charron† The mammalian genome contains two ERK/MAP kinase kinase genes, Map2k1 and Map2k2, which encode dual-specificity kinases responsible for ERK/MAP kinase activation. In the mouse, loss of Map2k1 function causes embryonic lethality, whereas Map2k2 mutants survive with a normal lifespan, suggesting that Map2k1 masks the phenotype due to the Map2k2 mutation. To uncover the specific function of MAP2K2 and the threshold requirement of MAP2K proteins during embryo formation, we have successively ablated the Map2k gene functions. We report here that Map2k2 haploinsufficiency affects the normal development of placenta in the absence of one Map2k1 allele. Most Map2k1+/–Map2k2+/– embryos die during gestation because of placenta defects restricted to extra-embryonic tissues. The impaired viability of Map2k1+/–Map2k2+/– embryos can be rescued when the Map2k1 deletion is restricted to the embryonic tissues. The severity of the placenta phenotype is dependent on the number of Map2k mutant alleles, the deletion of the Map2k1 allele being more deleterious. Moreover, the deletion of one or both Map2k2 alleles in the context of one null Map2k1 allele leads to the formation of multinucleated trophoblast giant (MTG) cells. Genetic experiments indicate that these structures are derived from Gcm1-expressing syncytiotrophoblasts (SynT), which are affected in their ability to form the uniform SynT layer II lining the maternal sinuses. Thus, even though Map2k1 plays a predominant role, these results enlighten the function of Map2k2 in placenta development. -

Supplementary Information Material and Methods
MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Supplementary Information Material and methods Chemicals The EGFR inhibitor NVP-AEE788 (Novartis), the Jak inhibitor I (Merck Calbiochem, #420099) and anisomycin (Alomone labs, # A-520) were prepared as 50 mM stock solutions in 100% DMSO. Doxorubicin (Adriablastin, Pfizer), EGF (Sigma Ref: E9644), PDGF (Sigma, Ref: P4306) and IL-4 (Sigma, Ref: I-4269) stock solutions were prepared as recommended by the manufacturer. For in vivo administration: Temodal (20 mg Temozolomide capsules, Essex Chemie AG, Luzern) was dissolved in 4 mL KZI/glucose (20/80, vol/vol); Taxotere was bought as 40 mg/mL solution (Sanofi Aventis, France), and prepared in KZI/glucose. Antibodies The primary antibodies used were as follows: anti-S473P-Akt (#9271), anti-T308P-Akt (#9276,), anti-S9P-GSK3β (#9336), anti-T389P-p70S6K (#9205), anti-YP/TP-Erk1/2 (#9101), anti-YP/TP-p38 (#9215), anti-YP/TP-JNK1/2 (#9101), anti-Y751P-PDGFR (#3161), anti- p21Cip1/Waf1 (#2946), anti-p27Kip1 (#2552) and anti-Ser15-p53 (#9284) antibodies were from Cell Signaling Technologies; anti-Akt (#05-591), anti-T32P-FKHRL1 (#06-952) and anti- PDGFR (#06-495) antibodies were from Upstate; anti-IGF-1R (#SC-713) and anti-EGFR (#SC-03) antibodies were from Santa Cruz; anti-GSK3α/β (#44610), anti-Y641P-Stat6 (#611566), anti-S1981P-ATM (#200-301), anti-T2609 DNA-PKcs (#GTX24194) and anti- 1 MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Y1316P-IGF-1R were from Bio-Source International, Becton-Dickinson, Rockland, GenTex and internal production, respectively. The 4G10 antibody was from Millipore (#05-321MG). -
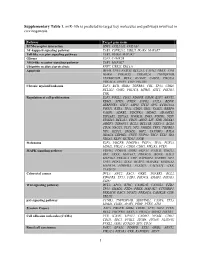
1 Supplementary Table 1. Mir-10B Is Predicted to Target Key Molecules
Supplementary Table 1. miR-10b is predicted to target key molecules and pathways involved in carcinogenesis. Pathway Target gene name ECM-receptor interaction SDC1, COL24A1, COL4A4 NF-kappa B signaling pathway TAB1, CSNK2A2, UBE2I, IRAK4, MAP3K7 Toll-like receptor signaling pathway TAB1, IRAK4, MAP3K7 Glioma E2F3, CAMK2B NOD-like receptor signaling pathway TAB1, MAP3K7 Ubiquitin mediated proteolysis RNF7, UBE2I, ERCC8 Apoptosis DFFB, TP53, FASLG, BCL2L1, CAPN2, PRKX, ATM, IRAK4, PRKACG, PRKAR2A, TNFRSF10B, TNFRSF10D, BCL2, IL1RAP, CASP8, PIK3CA, PRKACA, APAF1, CHP, PIK3R3 Chronic myeloid leukemia E2F3, BCR, GRB2, TGFBR1, CBL, TP53, CDK6, BCL2L1, GAB2, PIK3CA, MDM2, SHC1, PIK3R3, CRK Regulation of cell proliferation E2F3, FOSL2, CDX2, PDGFB, OSMR, E2F7, ARNT2, RBM5, STRN, PTEN, S1PR2, CUL3, BDNF, SERPINE1, SHC1, ASPH, ITCH, SPN, CCDC88A, FOXJ1, RXRA, TP53, CDK6, IRS1, VASH2, RBBP9, VASH1, ADRB2, PDGFRA, MDM2, ADAMTS1, EIF2AK2, EIF5A2, ICOSLG, ING5, FGFR3, NDN, ST8SIA1, BCL2L1, CDH5, ARNT, LIF, VDR, HOXA3, AGGF1, TSPAN31, BCL2, BCL11B, NKX3-1, BCL6, CD28, NACC1, FLT1, NF2, JARID2, TBX5, TGFBR1, NF1, KLF11, SMAD2, IGF2, TAX1BP3, BTLA, HDAC4, LEPRE1, CNTF, NUP62, TSC1, ETS1, ID4, NR5A2, KLF4, KCTD11, NFIB Melanoma E2F3, PDGFB, PDGFRA, FGF11, TP53, FGF23, MDM2, PIK3CA, CDK6, CDH1, PIK3R3, PTEN MAPK signaling pathway FGFR3, PDGFB, GRB2, FGF11, FASLG, GNG12, SRF, PRKX, MAP3K7, PRKACG, BDNF, RAC3, MAP3K2, PRKACA, CHP, RAPGEF2, TGFBR1, NF1, TP53, FGF23, STK4, DUSP5, MAP4K4, RPS6KA2, MAPK14, PDGFRA, PLA2G3, CACNA1C, CRK, PLA2G2F Colorectal cancer -

Hras Intracellular Trafficking and Signal Transduction Jodi Ho-Jung Mckay Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 HRas intracellular trafficking and signal transduction Jodi Ho-Jung McKay Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cancer Biology Commons, Cell Biology Commons, Genetics and Genomics Commons, and the Medical Cell Biology Commons Recommended Citation McKay, Jodi Ho-Jung, "HRas intracellular trafficking and signal transduction" (2007). Retrospective Theses and Dissertations. 13946. https://lib.dr.iastate.edu/rtd/13946 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. HRas intracellular trafficking and signal transduction by Jodi Ho-Jung McKay A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics Program of Study Committee: Janice E. Buss, Co-major Professor Linda Ambrosio, Co-major Professor Diane Bassham Drena Dobbs Ted Huiatt Iowa State University Ames, Iowa 2007 Copyright © Jodi Ho-Jung McKay, 2007. All rights reserved. UMI Number: 3274881 Copyright 2007 by McKay, Jodi Ho-Jung All rights reserved. UMI Microform 3274881 Copyright 2008 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. -

Predicting Coupling Probabilities of G-Protein Coupled Receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J
Published online 30 May 2019 Nucleic Acids Research, 2019, Vol. 47, Web Server issue W395–W401 doi: 10.1093/nar/gkz392 PRECOG: PREdicting COupling probabilities of G-protein coupled receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J. Silvio Gutkind4, Robert B. Russell1,2,* and Francesco Raimondi1,2,* 1CellNetworks, Bioquant, Heidelberg University, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany, 2Biochemie Zentrum Heidelberg (BZH), Heidelberg University, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany, 3Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai, Miyagi 980-8578, Japan and 4Department of Pharmacology and Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA Received February 10, 2019; Revised April 13, 2019; Editorial Decision April 24, 2019; Accepted May 01, 2019 ABSTRACT great use in tinkering with signalling pathways in living sys- tems (5). G-protein coupled receptors (GPCRs) control multi- Ligand binding to GPCRs induces conformational ple physiological states by transducing a multitude changes that lead to binding and activation of G-proteins of extracellular stimuli into the cell via coupling to situated on the inner cell membrane. Most of mammalian intra-cellular heterotrimeric G-proteins. Deciphering GPCRs couple with more than one G-protein giving each which G-proteins couple to each of the hundreds receptor a distinct coupling profile (6) and thus specific of GPCRs present in a typical eukaryotic organism downstream cellular responses. Determining these coupling is therefore critical to understand signalling. Here, profiles is critical to understand GPCR biology and phar- we present PRECOG (precog.russelllab.org): a web- macology. Despite decades of research and hundreds of ob- server for predicting GPCR coupling, which allows served interactions, coupling information is still missing for users to: (i) predict coupling probabilities for GPCRs many receptors and sequence determinants of coupling- specificity are still largely unknown. -

High Expression of LINC01268 Is Positively Associated with Hepatocellular Carcinoma Progression Via Regulating MAP3K7
OncoTargets and Therapy Dovepress open access to scientific and medical research Open Access Full Text Article ORIGINAL RESEARCH High Expression of LINC01268 is Positively Associated with Hepatocellular Carcinoma Progression via Regulating MAP3K7 This article was published in the following Dove Press journal: OncoTargets and Therapy Xiuli Jin,1 Weixin Fu,2 Dan Li,1 Objective: As one of the most common neoplastic diseases, hepatocellular carcinoma Ningning Wang,1 Jiayu Chen,1 (HCC) has a high morbidity and mortality, which seriously threatens human health and Zilu Zeng,1 Jiaqi Guo,1 Hao places a heavy burden on society and medical care. At present, effective early diagnosis, Liu,3 Xinping Zhong,3 Hu prognosis and treatment of HCC are limited. Altered gene expression patterns of lncRNA are Peng,4 Xin Yu,5 Jing Sun,1 associated with the occurrence, development and prognosis of various malignancies, includ- ing HCC. The aim of this study was to investigate the correlation between the expression of Xinhe Zhang,1 Xue Wang,1 LINC01268 and HCC, and to elucidate the potential underlying molecular mechanism. Beibei Xu,1 Yingbo Lin,6 4 Methods: Expression level and localization of LINC01268 in human liver cancer cells and Jianping Liu, Claudia HCC tissues were investigated using RT-qPCR and fluorescent in situ hybridization (FISH), 7 1 Kutter, Yiling Li respectively. Correlation of expression levels of LINC01268 and MAP3K7 with differentia- 1Department of Gastroenterology, First tion and poor overall patient survival of HCC were analyzed using in house collected and AffiliatedHospital of China Medical publicly available HCC tissue data. RT-qPCR and Western blot were applied to inspect the University, Shenyang, 110001, People’s Republic of China; 2Science Experiment effects of depletion and overexpression of LINC01268 on MAP3K7 expression.