Disintegration of the Scrophulariaceae1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Toward a Resolution of Campanulid Phylogeny, with Special Reference to the Placement of Dipsacales
TAXON 57 (1) • February 2008: 53–65 Winkworth & al. • Campanulid phylogeny MOLECULAR PHYLOGENETICS Toward a resolution of Campanulid phylogeny, with special reference to the placement of Dipsacales Richard C. Winkworth1,2, Johannes Lundberg3 & Michael J. Donoghue4 1 Departamento de Botânica, Instituto de Biociências, Universidade de São Paulo, Caixa Postal 11461–CEP 05422-970, São Paulo, SP, Brazil. [email protected] (author for correspondence) 2 Current address: School of Biology, Chemistry, and Environmental Sciences, University of the South Pacific, Private Bag, Laucala Campus, Suva, Fiji 3 Department of Phanerogamic Botany, The Swedish Museum of Natural History, Box 50007, 104 05 Stockholm, Sweden 4 Department of Ecology & Evolutionary Biology and Peabody Museum of Natural History, Yale University, P.O. Box 208106, New Haven, Connecticut 06520-8106, U.S.A. Broad-scale phylogenetic analyses of the angiosperms and of the Asteridae have failed to confidently resolve relationships among the major lineages of the campanulid Asteridae (i.e., the euasterid II of APG II, 2003). To address this problem we assembled presently available sequences for a core set of 50 taxa, representing the diver- sity of the four largest lineages (Apiales, Aquifoliales, Asterales, Dipsacales) as well as the smaller “unplaced” groups (e.g., Bruniaceae, Paracryphiaceae, Columelliaceae). We constructed four data matrices for phylogenetic analysis: a chloroplast coding matrix (atpB, matK, ndhF, rbcL), a chloroplast non-coding matrix (rps16 intron, trnT-F region, trnV-atpE IGS), a combined chloroplast dataset (all seven chloroplast regions), and a combined genome matrix (seven chloroplast regions plus 18S and 26S rDNA). Bayesian analyses of these datasets using mixed substitution models produced often well-resolved and supported trees. -

Central Appalachian Broadleaf Forest Coniferous Forest Meadow Province
Selecting Plants for Pollinators A Regional Guide for Farmers, Land Managers, and Gardeners In the Central Appalachian Broadleaf Forest Coniferous Forest Meadow Province Including the states of: Maryland, Pennsylvania, Virginia, West Virginia And parts of: Georgia, Kentucky, and North Carolina, NAPPC South Carolina, Tennessee Table of CONTENTS Why Support Pollinators? 4 Getting Started 5 Central Appalachian Broadleaf Forest 6 Meet the Pollinators 8 Plant Traits 10 Developing Plantings 12 Far ms 13 Public Lands 14 Home Landscapes 15 Bloom Periods 16 Plants That Attract Pollinators 18 Habitat Hints 20 This is one of several guides for Check list 22 different regions in the United States. We welcome your feedback to assist us in making the future Resources and Feedback 23 guides useful. Please contact us at [email protected] Cover: silver spotted skipper courtesy www.dangphoto.net 2 Selecting Plants for Pollinators Selecting Plants for Pollinators A Regional Guide for Farmers, Land Managers, and Gardeners In the Ecological Region of the Central Appalachian Broadleaf Forest Coniferous Forest Meadow Province Including the states of: Maryland, Pennsylvania, Virginia, West Virginia And parts of: Georgia, Kentucky, North Carolina, South Carolina, Tennessee a nappc and Pollinator Partnership™ Publication This guide was funded by the National Fish and Wildlife Foundation, the C.S. Fund, the Plant Conservation Alliance, the U.S. Forest Service, and the Bureau of Land Management with oversight by the Pollinator Partnership™ (www.pollinator.org), in support of the North American Pollinator Protection Campaign (NAPPC–www.nappc.org). Central Appalachian Broadleaf Forest – Coniferous Forest – Meadow Province 3 Why support pollinators? In theIr 1996 book, the Forgotten PollInators, Buchmann and Nabhan estimated that animal pollinators are needed for the reproduction “ Farming feeds of 90% of flowering plants and one third of human food crops. -

WHY SUCCULENT FLOWERS SEJ by A
Cape sugarbird WHY SUCCULENT FLOWERS SEJ by A. V. Milewski energy, water and nutrients which are The Field Guide to Trees of not retrieved by the plant, because the Southern Africa by Van Wyk & Van very purpose of fruit-pulp is to be Wyk (Struik 1997) shows the flowers removed from the plant once it is and fruits of trees and tall shrubs in ripe. The more resources the plant the south-western Cape. Of eighty spends on flowers, the less generous it species (excluding conifers) with is likely to be with fruit-pulp. Plants fleshy fruits eaten by birds, 91 % have may be generally faced with a choice small, dull flowers which easily of dispersing pollen or seeds, but not escape the notice of the human both, by avian flight. observer. Members of up to thirty-nine The division between bird-polli genera in twenty-five families conform nated and bird-sown plants is clear in in having fly-pollinated flowers and the south-western Cape, which has a bird-sown seeds in the south-western climate with winter rainfall and Cape. Eight percent (e.g. Glllysanthe summer drought similar to the maides, Grewia, Solanum) resemble lowers and fTllits vary greatly in Mediterranean Basin. Proteas charac many species of cultivated fruits size and how much food they teristic of this area produce large (e.g. apples, plums, raspberries) in Foffer polli nators. The smallest blooms pollinated by sunbirds and having colourful petals attracting flowers are dull greenish, and attract sugarbirds. Olives (Oleaceae) produce medium-size insects such as bees and pollinators no larger than small flies. -

Fair Use of This PDF File of Herbaceous
Fair Use of this PDF file of Herbaceous Perennials Production: A Guide from Propagation to Marketing, NRAES-93 By Leonard P. Perry Published by NRAES, July 1998 This PDF file is for viewing only. If a paper copy is needed, we encourage you to purchase a copy as described below. Be aware that practices, recommendations, and economic data may have changed since this book was published. Text can be copied. The book, authors, and NRAES should be acknowledged. Here is a sample acknowledgement: ----From Herbaceous Perennials Production: A Guide from Propagation to Marketing, NRAES- 93, by Leonard P. Perry, and published by NRAES (1998).---- No use of the PDF should diminish the marketability of the printed version. This PDF should not be used to make copies of the book for sale or distribution. If you have questions about fair use of this PDF, contact NRAES. Purchasing the Book You can purchase printed copies on NRAES’ secure web site, www.nraes.org, or by calling (607) 255-7654. Quantity discounts are available. NRAES PO Box 4557 Ithaca, NY 14852-4557 Phone: (607) 255-7654 Fax: (607) 254-8770 Email: [email protected] Web: www.nraes.org More information on NRAES is included at the end of this PDF. Acknowledgments This publication is an update and expansion of the 1987 Cornell Guidelines on Perennial Production. Informa- tion in chapter 3 was adapted from a presentation given in March 1996 by John Bartok, professor emeritus of agricultural engineering at the University of Connecticut, at the Connecticut Perennials Shortcourse, and from articles in the Connecticut Greenhouse Newsletter, a publication put out by the Department of Plant Science at the University of Connecticut. -

New York Non-Native Plant Invasiveness Ranking Form
NEW YORK NON -NATIVE PLANT INVASIVENESS RANKING FORM Scientific name: Callitriche stagnalis Scop. USDA Plants Code: CAST Common names: pond water-starwort Native distribution: Europe, Asia, North Africa Date assessed: July 1, 2008 Assessors: Steve Clemants Reviewers: LIISMA SRC Date Approved: July 9, 2008 Form version date: 10 July 2009 New York Invasiveness Rank: Low (Relative Maximum Score 40.00-49.99) Distribution and Invasiveness Rank (Obtain from PRISM invasiveness ranking form ) PRISM Status of this species in each PRISM: Current Distribution Invasiveness Rank 1 Adirondack Park Invasive Program Not Assessed Not Assessed 2 Capital/Mohawk Not Assessed Not Assessed 3 Catskill Regional Invasive Species Partnership Not Assessed Not Assessed 4 Finger Lakes Not Assessed Not Assessed 5 Long Island Invasive Species Management Area Restricted Insignificant 6 Lower Hudson Not Assessed Not Assessed 7 Saint Lawrence/Eastern Lake Ontario Not Assessed Not Assessed 8 Western New York Not Assessed Not Assessed Invasiveness Ranking Summary Total (Total Answered*) Total (see details under appropriate sub-section) Possible 1 Ecological impact 40 (30) 9 2 Biological characteristic and dispersal ability 25 (22 ) 12 3 Ecological amplitude and distribution 25 (25) 15 4 Difficulty of control 10 (3) 3 Outcome score 100 (80 )b 39a † Relative maximum score 48.75 § New York Invasiveness Rank Low (Relative Maximum Score 40.00-49.99) * For questions answered “unknown” do not include point value in “Total Answered Points Possible.” If “Total Answered Points Possible” is less than 70.00 points, then the overall invasive rank should be listed as “Unknown.” †Calculated as 100(a/b) to two decimal places. -

Well-Known Plants in Each Angiosperm Order
Well-known plants in each angiosperm order This list is generally from least evolved (most ancient) to most evolved (most modern). (I’m not sure if this applies for Eudicots; I’m listing them in the same order as APG II.) The first few plants are mostly primitive pond and aquarium plants. Next is Illicium (anise tree) from Austrobaileyales, then the magnoliids (Canellales thru Piperales), then monocots (Acorales through Zingiberales), and finally eudicots (Buxales through Dipsacales). The plants before the eudicots in this list are considered basal angiosperms. This list focuses only on angiosperms and does not look at earlier plants such as mosses, ferns, and conifers. Basal angiosperms – mostly aquatic plants Unplaced in order, placed in Amborellaceae family • Amborella trichopoda – one of the most ancient flowering plants Unplaced in order, placed in Nymphaeaceae family • Water lily • Cabomba (fanwort) • Brasenia (watershield) Ceratophyllales • Hornwort Austrobaileyales • Illicium (anise tree, star anise) Basal angiosperms - magnoliids Canellales • Drimys (winter's bark) • Tasmanian pepper Laurales • Bay laurel • Cinnamon • Avocado • Sassafras • Camphor tree • Calycanthus (sweetshrub, spicebush) • Lindera (spicebush, Benjamin bush) Magnoliales • Custard-apple • Pawpaw • guanábana (soursop) • Sugar-apple or sweetsop • Cherimoya • Magnolia • Tuliptree • Michelia • Nutmeg • Clove Piperales • Black pepper • Kava • Lizard’s tail • Aristolochia (birthwort, pipevine, Dutchman's pipe) • Asarum (wild ginger) Basal angiosperms - monocots Acorales -

Playing with Extremes Origins and Evolution of Exaggerated Female
Molecular Phylogenetics and Evolution 115 (2017) 95–105 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Playing with extremes: Origins and evolution of exaggerated female forelegs MARK in South African Rediviva bees ⁎ Belinda Kahnta,b, , Graham A. Montgomeryc, Elizabeth Murrayc, Michael Kuhlmannd,e, Anton Pauwf, Denis Michezg, Robert J. Paxtona,b, Bryan N. Danforthc a Institute of Biology/General Zoology, Martin-Luther-University Halle-Wittenberg, Hoher Weg 8, 06120 Halle (Saale), Germany b German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Deutscher Platz 5e, 04103 Leipzig, Germany c Department of Entomology, Cornell University, 3124 Comstock Hall, Ithaca, NY 14853-2601, USA d Zoological Museum, Kiel University, Hegewischstr. 3, 24105 Kiel, Germany e Dept. of Life Sciences, Natural History Museum, Cromwell Rd., London SW7 5BD, UK f Department of Botany and Zoology, Stellenbosch University, Matieland 7602, South Africa g Laboratoire de Zoologie, Research institute of Biosciences, University of Mons, Place du Parc 23, 7000 Mons, Belgium ARTICLE INFO ABSTRACT Keywords: Despite close ecological interactions between plants and their pollinators, only some highly specialised polli- Molecular phylogenetics nators adapt to a specific host plant trait by evolving a bizarre morphology. Here we investigated the evolution Plant-pollinator interaction of extremely elongated forelegs in females of the South African bee genus Rediviva (Hymenoptera: Melittidae), in Ecological adaptation which long forelegs are hypothesised to be an adaptation for collecting oils from the extended spurs of their Greater cape floristic region Diascia host flowers. We first reconstructed the phylogeny of the genus Rediviva using seven genes and inferred Trait evolution an origin of Rediviva at around 29 MYA (95% HPD = 19.2–40.5), concurrent with the origin and radiation of the Melittidae Succulent Karoo flora. -

Temporal and Spatial Origin of Gesneriaceae in the New World Inferred from Plastid DNA Sequences
bs_bs_banner Botanical Journal of the Linnean Society, 2013, 171, 61–79. With 3 figures Temporal and spatial origin of Gesneriaceae in the New World inferred from plastid DNA sequences MATHIEU PERRET1*, ALAIN CHAUTEMS1, ANDRÉA ONOFRE DE ARAUJO2 and NICOLAS SALAMIN3,4 1Conservatoire et Jardin botaniques de la Ville de Genève, Ch. de l’Impératrice 1, CH-1292 Chambésy, Switzerland 2Centro de Ciências Naturais e Humanas, Universidade Federal do ABC, Rua Santa Adélia, 166, Bairro Bangu, Santo André, Brazil 3Department of Ecology and Evolution, University of Lausanne, CH-1015 Lausanne, Switzerland 4Swiss Institute of Bioinformatics, Quartier Sorge, CH-1015 Lausanne, Switzerland Received 15 December 2011; revised 3 July 2012; accepted for publication 18 August 2012 Gesneriaceae are represented in the New World (NW) by a major clade (c. 1000 species) currently recognized as subfamily Gesnerioideae. Radiation of this group occurred in all biomes of tropical America and was accompanied by extensive phenotypic and ecological diversification. Here we performed phylogenetic analyses using DNA sequences from three plastid loci to reconstruct the evolutionary history of Gesnerioideae and to investigate its relationship with other lineages of Gesneriaceae and Lamiales. Our molecular data confirm the inclusion of the South Pacific Coronanthereae and the Old World (OW) monotypic genus Titanotrichum in Gesnerioideae and the sister-group relationship of this subfamily to the rest of the OW Gesneriaceae. Calceolariaceae and the NW genera Peltanthera and Sanango appeared successively sister to Gesneriaceae, whereas Cubitanthus, which has been previously assigned to Gesneriaceae, is shown to be related to Linderniaceae. Based on molecular dating and biogeographical reconstruction analyses, we suggest that ancestors of Gesneriaceae originated in South America during the Late Cretaceous. -
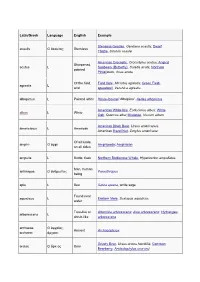
Dwarf Thistle, Cirsi
Latin/Greek Language English Example Stemless Gentian, Gentiana acaulis; Dwarf acaulis G ἄκαυλος Stemless Thistle, Cirsium acaule American Crocodile, Crocodylus acutus; Angled Sharpened, acutus L Sunbeam (Butterfly), Curetis acuta; Northern pointed Pintailduck, Anas acuta Of the field, Field Vole, Microtus agrestis; Green Field- agrestis L wild speedwell, Veronica agrestis albopictus L Painted white Hosta fortunei 'Albopicta', Aedes albopictus American White Ibis, Eudocimus albus; White albus L White Oak, Quercus alba; Mistletoe, Viscum album American Black Bear, Ursus americanus; americanus L American American Hazel Nut, Corylus americana Of all kinds, amphi- G ἀμφί Amphipoda; Amphibian on all sides ampulla L Bottle, flask Northern Bottlenose Whale, Hyperoodon ampullatus Man, human anthropos G ἄνθρωπος Paranthropus being apis L Bee Salvia apiana, white sage Found near aquaticus L Eastern Mole, Scalopus aquaticus water Tree-like or Artemisia arborescens; Aloe arborescens; Hydrangea arborescens L shrub-like arborescens archaeos, G ἀρχαῖος, Ancient Archaeopteryx archaeo- ἀρχαιο- Grizzly Bear, Ursus arctos horribilis; Common arctos G ἄρκτος Bear Bearberry, Arctostaphylos uva-ursi argentatus L Silvery Herring Gull, Larus argentatus arthron G ἄρθρον Joint Arthropoda arvensis L In the field Skylark, Alauda arvensis astron, astro-, G ἄστρον, Star Starfish (class), Asteroidea astero- ἀστρο-, ἀστερο- Acer palmatum 'Atropurpureum'; Berberis atropurpureum L Deep purple thunbergii f. atropurpurea Daphne odora 'Aureomarginata'; Taxus aureomarginata -

Diagnosis and Differentiation of the Order Primates
YEARBOOK OF PHYSICAL ANTHROPOLOGY 30:75-105 (1987) Diagnosis and Differentiation of the Order Primates FREDERICK S. SZALAY, ALFRED L. ROSENBERGER, AND MARIAN DAGOSTO Department of Anthropolog* Hunter College, City University of New York, New York, New York 10021 (F.S.S.); University of Illinois, Urbanq Illinois 61801 (A.L. R.1; School of Medicine, Johns Hopkins University/ Baltimore, h4D 21218 (M.B.) KEY WORDS Semiorders Paromomyiformes and Euprimates, Suborders Strepsirhini and Haplorhini, Semisuborder Anthropoidea, Cranioskeletal morphology, Adapidae, Omomyidae, Grades vs. monophyletic (paraphyletic or holophyletic) taxa ABSTRACT We contrast our approach to a phylogenetic diagnosis of the order Primates, and its various supraspecific taxa, with definitional proce- dures. The order, which we divide into the semiorders Paromomyiformes and Euprimates, is clearly diagnosable on the basis of well-corroborated informa- tion from the fossil record. Lists of derived features which we hypothesize to have been fixed in the first representative species of the Primates, Eupri- mates, Strepsirhini, Haplorhini, and Anthropoidea, are presented. Our clas- sification of the order includes both holophyletic and paraphyletic groups, depending on the nature of the available evidence. We discuss in detail the problematic evidence of the basicranium in Paleo- gene primates and present new evidence for the resolution of previously controversial interpretations. We renew and expand our emphasis on postcra- nial analysis of fossil and living primates to show the importance of under- standing their evolutionary morphology and subsequent to this their use for understanding taxon phylogeny. We reject the much advocated %ladograms first, phylogeny next, and scenario third” approach which maintains that biologically founded character analysis, i.e., functional-adaptive analysis and paleontology, is irrelevant to genealogy hypotheses. -

A Revised Taxonomy of the Iguanodont Dinosaur Genera and Species
ARTICLE IN PRESS + MODEL Cretaceous Research xx (2007) 1e25 www.elsevier.com/locate/CretRes A revised taxonomy of the iguanodont dinosaur genera and species Gregory S. Paul 3109 North Calvert Station, Side Apartment, Baltimore, MD 21218-3807, USA Received 20 April 2006; accepted in revised form 27 April 2007 Abstract Criteria for designating dinosaur genera are inconsistent; some very similar species are highly split at the generic level, other anatomically disparate species are united at the same rank. Since the mid-1800s the classic genus Iguanodon has become a taxonomic grab-bag containing species spanning most of the Early Cretaceous of the northern hemisphere. Recently the genus was radically redesignated when the type was shifted from nondiagnostic English Valanginian teeth to a complete skull and skeleton of the heavily built, semi-quadrupedal I. bernissartensis from much younger Belgian sediments, even though the latter is very different in form from the gracile skeletal remains described by Mantell. Currently, iguanodont remains from Europe are usually assigned to either robust I. bernissartensis or gracile I. atherfieldensis, regardless of lo- cation or stage. A stratigraphic analysis is combined with a character census that shows the European iguanodonts are markedly more morpho- logically divergent than other dinosaur genera, and some appear phylogenetically more derived than others. Two new genera and a new species have been or are named for the gracile iguanodonts of the Wealden Supergroup; strongly bipedal Mantellisaurus atherfieldensis Paul (2006. Turning the old into the new: a separate genus for the gracile iguanodont from the Wealden of England. In: Carpenter, K. (Ed.), Horns and Beaks: Ceratopsian and Ornithopod Dinosaurs. -

Towards Resolving Lamiales Relationships
Schäferhoff et al. BMC Evolutionary Biology 2010, 10:352 http://www.biomedcentral.com/1471-2148/10/352 RESEARCH ARTICLE Open Access Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences Bastian Schäferhoff1*, Andreas Fleischmann2, Eberhard Fischer3, Dirk C Albach4, Thomas Borsch5, Günther Heubl2, Kai F Müller1 Abstract Background: In the large angiosperm order Lamiales, a diverse array of highly specialized life strategies such as carnivory, parasitism, epiphytism, and desiccation tolerance occur, and some lineages possess drastically accelerated DNA substitutional rates or miniaturized genomes. However, understanding the evolution of these phenomena in the order, and clarifying borders of and relationships among lamialean families, has been hindered by largely unresolved trees in the past. Results: Our analysis of the rapidly evolving trnK/matK, trnL-F and rps16 chloroplast regions enabled us to infer more precise phylogenetic hypotheses for the Lamiales. Relationships among the nine first-branching families in the Lamiales tree are now resolved with very strong support. Subsequent to Plocospermataceae, a clade consisting of Carlemanniaceae plus Oleaceae branches, followed by Tetrachondraceae and a newly inferred clade composed of Gesneriaceae plus Calceolariaceae, which is also supported by morphological characters. Plantaginaceae (incl. Gratioleae) and Scrophulariaceae are well separated in the backbone grade; Lamiaceae and Verbenaceae appear in distant clades, while the recently described Linderniaceae are confirmed to be monophyletic and in an isolated position. Conclusions: Confidence about deep nodes of the Lamiales tree is an important step towards understanding the evolutionary diversification of a major clade of flowering plants. The degree of resolution obtained here now provides a first opportunity to discuss the evolution of morphological and biochemical traits in Lamiales.