Metabolic Stress Promotes Stop-Codon Readthrough and Phenotypic Heterogeneity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Crystal Structure of a Translation Termination Complex Formed with Release Factor RF2
Crystal structure of a translation termination complex formed with release factor RF2 Andrei Korosteleva,b,1, Haruichi Asaharaa,b,1,2, Laura Lancastera,b,1, Martin Laurberga,b, Alexander Hirschia,b, Jianyu Zhua,b, Sergei Trakhanova,b, William G. Scotta,c, and Harry F. Nollera,b,3 aCenter for Molecular Biology of RNA and Departments of bMolecular, Cell and Developmental Biology and cChemistry and Biochemistry, University of California, Santa Cruz, CA 95064 Contributed by Harry F. Noller, October 30, 2008 (sent for review October 22, 2008) We report the crystal structure of a translation termination com- and G530 of 16S rRNA, and recognized separately by interac- plex formed by the Thermus thermophilus 70S ribosome bound tions with Gln-181 and Thr-194. Stop codon recognition by RF1 with release factor RF2, in response to a UAA stop codon, solved also involves a network of interactions with other structural at 3 Å resolution. The backbone of helix ␣5 and the side chain of elements of RF1, including critical main-chain atoms and con- serine of the conserved SPF motif of RF2 recognize U1 and A2 of the served features of 16S rRNA (9). stop codon, respectively. A3 is unstacked from the first 2 bases, Many studies have implicated the conserved GGQ motif in contacting Thr-216 and Val-203 of RF2 and stacking on G530 of 16S domain 3, present in the release factors of all three primary rRNA. The structure of the RF2 complex supports our previous domains of life, in the hydrolysis reaction. Although the side proposal that conformational changes in the ribosome in response chain of the conserved glutamine has been proposed to play a to recognition of the stop codon stabilize rearrangement of the role in catalysis (10, 11), elimination of its side-chain amide switch loop of the release factor, resulting in docking of the group by mutation of this residue to alanine, for example, universally conserved GGQ motif in the PTC of the 50S subunit. -

Structural Aspects of Translation Termination on the Ribosome
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by eScholarship@UMMS University of Massachusetts Medical School eScholarship@UMMS RNA Therapeutics Institute Publications RNA Therapeutics Institute 2011-08-01 Structural aspects of translation termination on the ribosome Andrei A. Korostelev University of Massachusetts Medical School Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/rti_pubs Part of the Biochemistry, Biophysics, and Structural Biology Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, and the Therapeutics Commons Repository Citation Korostelev AA. (2011). Structural aspects of translation termination on the ribosome. RNA Therapeutics Institute Publications. https://doi.org/10.1261/rna.2733411. Retrieved from https://escholarship.umassmed.edu/rti_pubs/33 This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in RNA Therapeutics Institute Publications by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. REVIEW Structural aspects of translation termination on the ribosome ANDREI A. KOROSTELEV1 RNA Therapeutics Institute and Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, Worcester, Massachusetts 01605, USA ABSTRACT Translation of genetic information encoded in messenger RNAs into polypeptide sequences is carried out by ribosomes in all organisms. When a full protein is synthesized, a stop codon positioned in the ribosomal A site signals termination of translation and protein release. Translation termination depends on class I release factors. Recently, atomic-resolution crystal structures were determined for bacterial 70S ribosome termination complexes bound with release factors RF1 or RF2. -

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -

Bio 102 Practice Problems Genetic Code and Mutation
Bio 102 Practice Problems Genetic Code and Mutation Multiple choice: Unless otherwise directed, circle the one best answer: 1. Choose the one best answer: Beadle and Tatum mutagenized Neurospora to find strains that required arginine to live. Based on the classification of their mutants, they concluded that: A. one gene corresponds to one protein. B. DNA is the genetic material. C. "inborn errors of metabolism" were responsible for many diseases. D. DNA replication is semi-conservative. E. protein cannot be the genetic material. 2. Choose the one best answer. Which one of the following is NOT part of the definition of a gene? A. A physical unit of heredity B. Encodes a protein C. Segement of a chromosome D. Responsible for an inherited characteristic E. May be linked to other genes 3. A mutation converts an AGA codon to a TGA codon (in DNA). This mutation is a: A. Termination mutation B. Missense mutation C. Frameshift mutation D. Nonsense mutation E. Non-coding mutation 4. Beadle and Tatum performed a series of complex experiments that led to the idea that one gene encodes one enzyme. Which one of the following statements does not describe their experiments? A. They deduced the metabolic pathway for the synthesis of an amino acid. B. Many different auxotrophic mutants of Neurospora were isolated. C. Cells unable to make arginine cannot survive on minimal media. D. Some mutant cells could survive on minimal media if they were provided with citrulline or ornithine. E. Homogentisic acid accumulates and is excreted in the urine of diseased individuals. 5. -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -
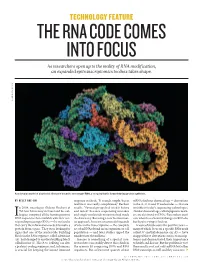
THE RNA CODE COMES INTO FOCUS As Researchers Open up to the Reality of RNA Modification, an Expanded Epitranscriptomics Toolbox Takes Shape
TECHNOLOGY FEATURE THE RNA CODE COMES INTO FOCUS As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape. LAGUNA DESIGN/SPL LAGUNA A molecular model of a bacterial ribosome bound to messenger RNA, a complex that is formed during protein synthesis. BY KELLY RAE CHI response in check. “It sounds simple, but in mRNAs harbour chemical tags — decorations real life it was really complicated,” Rechavi to the A, C, G and U nucleotides — that are n 2004, oncologist Gideon Rechavi at recalls. “Several groups had tried it before invisible to today’s sequencing technologies. Tel Aviv University in Israel and his col- and failed” because sequencing mistakes (Similar chemical tags, called epigenetic mark- leagues compared all the human genomic and single-nucleotide mutations had made ers, are also found on DNA.) Researchers aren’t IDNA sequences then available with their cor- the data noisy. But using a new bioinformat- sure what these chemical changes in RNA do, responding messenger RNAs — the molecules ics approach, his team uncovered thousands but they’re trying to find out. that carry the information needed to make a of sites in the transcriptome — the complete A wave of studies over the past five years — protein from a gene. They were looking for set of mRNAs found in an organism or cell many of which focus on a specific RNA mark signs that one of the nucleotide building population — and later studies upped the called N6-methyladenosine (m6A) — have blocks in the RNA sequence, called adenosine number into the millions1. -

Translational Control Using an Expanded Genetic Code
Review Translational Control using an Expanded Genetic Code Yusuke Kato Division of Biotechnology, Institute of Agrobiological Sciences, National Agriculture and Food Research Organization (NARO), Oowashi 1-2, Tsukuba, Ibaraki 305-8634, Japan; [email protected]; Tel.: +81-29-838-6059 Received: 19 December 2018; Accepted: 15 February 2019; Published: 18 February 2019 Abstract: A bio-orthogonal and unnatural substance, such as an unnatural amino acid (Uaa), is an ideal regulator to control target gene expression in a synthetic gene circuit. Genetic code expansion technology has achieved Uaa incorporation into ribosomal synthesized proteins in vivo at specific sites designated by UAG stop codons. This site-specific Uaa incorporation can be used as a controller of target gene expression at the translational level by conditional read-through of internal UAG stop codons. Recent advances in optimization of site-specific Uaa incorporation for translational regulation have enabled more precise control over a wide range of novel important applications, such as Uaa-auxotrophy-based biological containment, live-attenuated vaccine, and high-yield zero-leakage expression systems, in which Uaa translational control is exclusively used as an essential genetic element. This review summarizes the history and recent advance of the translational control by conditional stop codon read-through, especially focusing on the methods using the site-specific Uaa incorporation. Keywords: genetic switch; synthetic biology; unnatural amino acids; translational regulation; biological containment; stop codon read-through 1. Introduction Conditional induction of gene expression is one of the most important technologies for constructing synthetic gene circuits, and such regulation has applications from basic research to industrial use. -

Ef-G:Trna Dynamics During the Elongation Cycle of Protein Synthesis
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2015 Ef-G:trna Dynamics During the Elongation Cycle of Protein Synthesis Rong Shen University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Biochemistry Commons Recommended Citation Shen, Rong, "Ef-G:trna Dynamics During the Elongation Cycle of Protein Synthesis" (2015). Publicly Accessible Penn Dissertations. 1131. https://repository.upenn.edu/edissertations/1131 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1131 For more information, please contact [email protected]. Ef-G:trna Dynamics During the Elongation Cycle of Protein Synthesis Abstract During polypeptide elongation cycle, prokaryotic elongation factor G (EF-G) catalyzes the coupled translocations on the ribosome of mRNA and A- and P-site bound tRNAs. Continued progress has been achieved in understanding this key process, including results of structural, ensemble kinetic and single- molecule studies. However, most of work has been focused on the pre-equilibrium states of this fast process, leaving the real time dynamics, especially how EF-G interacts with the A-site tRNA in the pretranslocation complex, not fully elucidated. In this thesis, the kinetics of EF-G catalyzed translocation is investigated by both ensemble and single molecule fluorescence resonance energy transfer studies to further explore the underlying mechanism. In the ensemble work, EF-G mutants were designed and expressed successfully. The labeled EF-G mutants show good translocation activity in two different assays. In the smFRET work, by attachment of a fluorescent probe at position 693 on EF-G permits monitoring of FRET efficiencies to sites in both ribosomal protein L11 and A-site tRNA. -

Mrna, Rrna and Trna Types of RNA: Mrna, Rrna and Trna
Types of RNA: mRNA, rRNA and tRNA Types of RNA: mRNA, rRNA and tRNA By Susha Cheriyedath, M.Sc. Reviewed by Michael Greenwood, M.Sc. RNA or ribonucleic acid is a polymer of nucleotides that is made up of a ribose sugar, a phosphate, and bases such as adenine, guanine, cytosine, and uracil. It plays a crucial role in gene expression by acting as the intermediate between the genetic information encoded by DNA and proteins. Designua | Shutterstock RNA has a structure very similar to that of DNA. The key difference in RNA structure is that the ribose sugar in RNA possesses a hydroxyl (OH) group that is absent in DNA. Types of RNA In both prokaryotes and eukaryotes, there are three main types of RNA – messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). These 3 types of RNA are discussed below. P Saved from URL: https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx 1/5 Types of RNA: mRNA, rRNA and tRNA Messenger RNA (mRNA) mRNA accounts for just 5% of the total RNA in the cell. mRNA is the most heterogeneous of the 3 types of RNA in terms of both base sequence and size. It carries complimentary genetic code copied, from DNA during transcription, in the form of triplets of nucleotides called codons. Each codon specifies a particular amino acid, though one amino acid may be coded for by many different codons. Although there are 64 possible codons or triplet bases in the genetic code, only 20 of them represent amino acids. -

Transcription Study Guide This Study Guide Is a Written Version of the Material You Have Seen Presented in the Transcription Unit
Transcription Study Guide This study guide is a written version of the material you have seen presented in the transcription unit. The cell’s DNA contains the instructions for carrying out the work of the cell. These instructions are used by the cell’s protein-making machinery to create proteins. If the cell’s DNA were directly read by the protein-making machinery, however, it could be damaged and the process would be slow and cumbersome. The cell avoids this problem by copying genetic information from its DNA into an intermediate called messenger RNA (mRNA). It is this mRNA that is read by the cell’s protein-making machinery. This process is called transcription. Components In this section you will be introduced to the components involved in the process of RNA synthesis, called transcription. This process requires an enzyme that uses many nucleotide bases to copy the instructions present in DNA into an intermediate messenger RNA molecule. RNA What is RNA? · Like DNA, RNA is a polymer made up of nucleotides. · Unlike DNA, which is composed of two strands of nucleotides twisted together, RNA is single-stranded. It can also sometimes fold into complex three-dimensional structures. · RNA contains the same nucleotides as DNA, with the substitution of uraciluridine (U) for thymidine (T). · RNA is chemically different from DNA so that the cell can easily tell the two apart. · In this animation, you will see one type of RNA, messenger RNA, being put together. · There are three types of RNA: mRNA, which you will read more about; tRNA, which is used in the translation process, and rRNA, which acts as a structural element in the ribosome (a translation component). -

Isolation of RNA with Properties of Messenger RNA from Cerebral Polyribosomes* Claire E
Proceedings of the National Academy of Sciences Vol. 67, No. 2, pp. 644-651, October 1970 Isolation of RNA with Properties of Messenger RNA from Cerebral Polyribosomes* Claire E. Zomzelyt, Sidney Roberts, and Susan Peache DEPARTMENT OF BIOLOGICAL CHEMISTRY, SCHOOL OF MEDICINE, AND THE BRAIN RESEARCH INSTI- TUTE, UNIVERSITY OF CALIFORNIA CENTER FOR THE HEALTH SCIENCES, LOS ANGELES, CALIFORNIA 90024 Communicated by H. W. Magoun, July 24, 1970 Abstract. RNA which dissociated from purified cerebral polyribosomes of adult rats in the presence of EDTA was isolated by fractionation in a dis- continuous sucrose gradient. The yield was 2% of the total polyribosomal RNA. The base composition resembled the complementary values for rat DNA and was very different from base compositions of ribosomal RNA and transfer RNA. This RNA fraction contained a large proportion of molecules which were rapidly labeled in vivo and hybridized to homologous DNA. The polyribosomal RNA preparation also exhibited high template activity in a cerebral cell-free system which had previously been stripped of the capacity to incorporate amino acids in the absence of added messenger RNA (mRNA). Sedimentation analysis revealed only two peaks, with coefficients of approximately 8 S and 16 S. The data indicate that RNA with the properties of mRNA can be selectively isolated from cerebral polyribosomes under mild conditions which avoid degradation. Numerous studies support the concept that alterations in RNA and protein synthesis are associated with information transfer and storage in the central nervous system. Activation of the synthesis of specific proteins is suggested by the reports that new RNA with altered base composition, presumably mRNA, is formed during the acquisition phase of learning.