Global Alterations to the Choroid Plexus Blood-CSF Barrier in Amyotrophic Lateral Sclerosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Local Genetic and Environmental Factors in Asthma Disease Pathogenesis: Chronicity and Persistence Mechanisms
Eur Respir J 2007; 29: 793–803 DOI: 10.1183/09031936.00087506 CopyrightßERS Journals Ltd 2007 SERIES ‘‘GENETICS OF ASTHMA AND COPD IN THE POSTGENOME ERA’’ Edited by E. von Mutius, M. Kabesch and F. Kauffmann Number 4 in this Series Local genetic and environmental factors in asthma disease pathogenesis: chronicity and persistence mechanisms S.T. Holgate*, D.E. Davies*, R.M. Powell*, P.H. Howarth*, H.M. Haitchi# and J.W. Holloway" ABSTRACT: While asthma is an inflammatory disorder of the airways usually associated with AFFILIATIONS atopy, an important additional component is involvement of the epithelium and underlying *Allergy and Inflammation Research, Division of Infection, Inflammation mesenchyme acting as a trophic unit (EMTU). and Repair, In addition to allergens, a wide range of environmental factors interact with the EMTU, such as #IIR Division and virus infections, environmental tobacco smoke and pollutants, to initiate tissue damage and "Division of Human Genetics, School aberrant repair responses that are translated into remodelling of the airways. While candidate of Medicine, Southampton General Hospital, Southampton, UK. gene association studies have revealed polymorphic variants that influence asthmatic inflamma- tion, positional cloning of previously unknown genes is identifying a high proportion of novel CORRESPONDENCE genes in the EMTU. S.T. Holgate Dipeptidyl peptidase (DPP) 10 and disintegrin and metalloproteinase (ADAM)33 are newly Allergy and Inflammation Research MP810 identified genes strongly associated with asthma that are preferentially expressed in the airway Level D epithelium and underlying mesenchyme, respectively. Centre Block Also of increasing importance is the recognition that genes associated with asthma and atopy Southampton General Hospital have important interactions with the environment through epigenetic mechanisms that influence Southampton SO16 6YD UK their expression. -

PCDH1 Antibody (Monoclonal) (M01) Mouse Monoclonal Antibody Raised Against a Partial Recombinant PCDH1
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 PCDH1 Antibody (monoclonal) (M01) Mouse monoclonal antibody raised against a partial recombinant PCDH1. Catalog # AT3210a Specification PCDH1 Antibody (monoclonal) (M01) - Product Information Application IF, WB, IHC, E Primary Accession Q08174 Other Accession NM_002587 Reactivity Human Host mouse Clonality Monoclonal Isotype IgG1 Kappa Calculated MW 114743 PCDH1 Antibody (monoclonal) (M01) - Additional Information Immunofluorescence of monoclonal antibody to PCDH1 on A-431 cell. [antibody concentration 10 ug/ml] Gene ID 5097 Other Names Protocadherin-1, Cadherin-like protein 1, Protocadherin-42, PC42, PCDH1 Target/Specificity PCDH1 (NP_002578, 62 a.a. ~ 169 a.a) partial recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Dilution WB~~1:500~1000 Format Clear, colorless solution in phosphate Antibody Reactive Against Recombinant buffered saline, pH 7.2 . Protein.Western Blot detection against Immunogen (37.62 KDa) . Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Precautions PCDH1 Antibody (monoclonal) (M01) is for research use only and not for use in diagnostic or therapeutic procedures. PCDH1 Antibody (monoclonal) (M01) - Protocols Page 1/3 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Provided below are standard protocols that you may find useful for product applications. • Western Blot • Blocking Peptides • Dot Blot • Immunohistochemistry • Immunofluorescence • Immunoprecipitation • Flow Cytomety • Cell Culture PCDH1 monoclonal antibody (M01), clone 5D5 Western Blot analysis of PCDH1 expression in A-431 ( (Cat # AT3210a ) Immunoperoxidase of monoclonal antibody to PCDH1 on formalin-fixed paraffin-embedded human salivary gland. -

Global Alterations to the Choroid Plexus Blood-CSF Barrier in Amyotrophic Lateral Sclerosis J
Saul et al. Acta Neuropathologica Communications (2020) 8:92 https://doi.org/10.1186/s40478-020-00968-9 RESEARCH Open Access Global alterations to the choroid plexus blood-CSF barrier in amyotrophic lateral sclerosis J. Saul1,2, E. Hutchins3, R. Reiman3, M. Saul4, L. W. Ostrow5, B. T. Harris6, K. Van Keuren-Jensen3, R. Bowser1,2 and N. Bakkar1,2* Abstract The choroid plexus (CP) is a highly vascularized structure located in the ventricles that forms the blood-CSF barrier (BCSFB) and separates the blood from the cerebrospinal fluid (CSF). In addition to its role as a physical barrier, the CP functions in CSF secretion, transport of nutrients into the central nervous system (CNS) and a gated point of entry of circulating immune cells into the CNS. Aging and neurodegeneration have been reported to affect CP morphology and function and increase protein leakage from blood to the CSF. Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease associated with both upper and lower motor neuron loss, as well as altered proteomic and metabolomic signatures in the CSF. The role of the BCSFB and the CP in ALS is unknown. Here we describe a transcriptomic and ultrastructural analysis of BCSFB and CP alterations in human postmortem tissues from ALS and non-neurologic disease controls. ALS-CP exhibited widespread disruptions in tight junctional components of the CP epithelial layer and vascular integrity. In addition, we detected loss of pericytes around ALS blood vessels, accompanied by activation of platelet aggregation markers vWF and Fibrinogen, reminiscent of vascular injury. To investigate the immune component of ALS-CP, we conducted a comprehensive analysis of cytokines and chemokine panels in CP lysates and found a significant down-regulation of M- CSF and V-CAM1 in ALS, as well as up-regulation of VEGF-A protein. -

Cell Adhesion Molecules in Normal Skin and Melanoma
biomolecules Review Cell Adhesion Molecules in Normal Skin and Melanoma Cian D’Arcy and Christina Kiel * Systems Biology Ireland & UCD Charles Institute of Dermatology, School of Medicine, University College Dublin, D04 V1W8 Dublin, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +353-1-716-6344 Abstract: Cell adhesion molecules (CAMs) of the cadherin, integrin, immunoglobulin, and selectin protein families are indispensable for the formation and maintenance of multicellular tissues, espe- cially epithelia. In the epidermis, they are involved in cell–cell contacts and in cellular interactions with the extracellular matrix (ECM), thereby contributing to the structural integrity and barrier for- mation of the skin. Bulk and single cell RNA sequencing data show that >170 CAMs are expressed in the healthy human skin, with high expression levels in melanocytes, keratinocytes, endothelial, and smooth muscle cells. Alterations in expression levels of CAMs are involved in melanoma propagation, interaction with the microenvironment, and metastasis. Recent mechanistic analyses together with protein and gene expression data provide a better picture of the role of CAMs in the context of skin physiology and melanoma. Here, we review progress in the field and discuss molecular mechanisms in light of gene expression profiles, including recent single cell RNA expression information. We highlight key adhesion molecules in melanoma, which can guide the identification of pathways and Citation: D’Arcy, C.; Kiel, C. Cell strategies for novel anti-melanoma therapies. Adhesion Molecules in Normal Skin and Melanoma. Biomolecules 2021, 11, Keywords: cadherins; GTEx consortium; Human Protein Atlas; integrins; melanocytes; single cell 1213. https://doi.org/10.3390/ RNA sequencing; selectins; tumour microenvironment biom11081213 Academic Editor: Sang-Han Lee 1. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

(12) Patent Application Publication (10) Pub. No.: US 2005/0260639 A1 Nakamura Et Al
US 2005O260639A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2005/0260639 A1 Nakamura et al. (43) Pub. Date: Nov. 24, 2005 (54) METHOD FOR DIAGNOSING PANCREATIC Related U.S. Application Data CANCER (63) Continuation-in-part of application No. PCT/JPO3/ 11817, filed on Sep. 17, 2003. (75) Inventors: Yusuke Nakamura, Yokohama-shi (JP); Toyomasa Katagiri, Shinagawa-ku (60) Provisional application No. 60/555,809, filed on Mar. 24, 2004. Provisional application No. 60/450,889, (JP); Hidewaki Nakagawa, filed on Feb. 28, 2003. Provisional application No. Shinagawa-ku (JP) 60/414,872, filed on Sep. 30, 2002. Publication Classification Correspondence Address: TOWNSEND AND TOWNSEND AND CREW, (51) Int. Cl. .................................................. C12O 1/68 LLP (52) U.S. Cl. .................................................................. 435/6 TWO EMBARCADERO CENTER EIGHTH FLOOR (57) ABSTRACT SAN FRANCISCO, CA 94111-3834 (US) Objective methods for detecting and diagnosing pancreatic cancer (PNC) are described herein. In one embodiment, the (73) Assignees: Oncotherapy Science, Inc., Kawasaki diagnostic method involves determining the expression level shi (JP); The University of Tokyo, Bun of PNC-associated gene that discriminates between PNC kyo-ku (JP) cells and normal cells. The present invention further pro vides methods of Screening for therapeutic agents useful in (21) Appl. No.: 11/090,739 the treatment of pancreatic cancer, methods of treating pancreatic cancer and method of vaccinating a Subject (22) Filed: Mar. 24, 2005 against pancreatic cancer. Patent Application Publication Nov. 24, 2005 Sheet 1 of 16 US 2005/0260639 A1 FG 1 Patent Application Publication Nov. 24, 2005 Sheet 2 of 16 US 2005/0260639 A1 s - to co on S 2 2 O C C C D C C C D C) () () O U O n. -
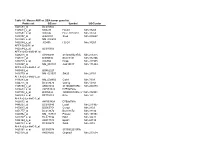
Table S1: Mouse AKR Vs DBA Tumor Gene List Probe Set GB Acc Symbol
Table S1: Mouse AKR vs DBA tumor gene list Probe set GB acc Symbol UGCluster 1425583_at BC010605 1425614_x_at M83244 H2-D1 Mm.33263 1427651_x_at X00246 H2-L /// H2-D1 Mm.33263 1419157_at AI428101 Sox4 Mm.240627 1422565_s_at NM_008688 1452544_x_at J00406 H2-D1 Mm.33263 AFFX-BioB-M_at 1425584_x_at BC010605 AFFX-r2-Ec-bioB-M_at 1426278_at AY090098 2310061N23Rik Mm.271275 1423411_at BI099836 BC013481 Mm.332406 1459725_s_at C86550 Dcpp Mm.287985 1419327_at NM_053181 AA415817 Mm.171484 AFFX-r2-Ec-bioB-3_at 1455869_at BG862223 1416770_at NM_021537 Stk25 Mm.28761 AFFX-r2-Ec-bioB-5_at 1418283_at NM_009903 Cldn4 Mm.7339 1424775_at BC018470 Oas1g Mm.14301 1428850_x_at AK004342 2410026K10Rik Mm.260878 1426633_s_at AW553424 D7Ertd760e 1427932_s_at BI076834 1200003I10Rik /// 1200015M12RikMm.332931 /// 1200016E24Rik 1449289_a_at BF715219 B2m Mm.163 AFFX-r2-Ec-bioC-3_at 1426632_at AW553424 D7Ertd760e 1448207_at BC010840 Lasp1 Mm.271967 1450017_at BG065754 Ccng1 Mm.2103 1451777_at BC013672 BC013672 Mm.33332 1420352_at NM_133731 Prss22 Mm.157351 1423747_a_at BC027196 Pdk1 Mm.34411 1454169_a_at AK017174 Epsti1 Mm.68134 1448793_a_at BC005679 Sdc4 Mm.3815 AFFX-r2-Ec-bioC-5_at 1425161_a_at BC005574 5730502D15Rik 1423158_at AK008566 Gnpnat1 Mm.233534 1453196_a_at BQ033138 Oasl2 Mm.228363 1449250_at NM_033573 Prcc Mm.35089 1452428_a_at AI099111 B2m Mm.163 1421024_at BB524140 Agpat1 Mm.8684 1450016_at BG065754 Ccng1 Mm.2103 1419043_a_at BM239828 AW111922 Mm.326506 1449262_s_at BB704337 Lin7c Mm.235300 1426975_at BG067859 4632413K17Rik Mm.295246 1426164_a_at AF479773 -

Targeting the DEK Oncogene in Head and Neck Squamous Cell Carcinoma: Functional and Transcriptional Consequences
Targeting the DEK oncogene in head and neck squamous cell carcinoma: functional and transcriptional consequences A dissertation submitted to the Graduate School of the University of Cincinnati in partial fulfillment of the requirements to the degree of Doctor of Philosophy (Ph.D.) in the Department of Cancer and Cell Biology of the College of Medicine March 2015 by Allie Kate Adams B.S. The Ohio State University, 2009 Dissertation Committee: Susanne I. Wells, Ph.D. (Chair) Keith A. Casper, M.D. Peter J. Stambrook, Ph.D. Ronald R. Waclaw, Ph.D. Susan E. Waltz, Ph.D. Kathryn A. Wikenheiser-Brokamp, M.D., Ph.D. Abstract Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignancies worldwide with over 50,000 new cases in the United States each year. For many years tobacco and alcohol use were the main etiological factors; however, it is now widely accepted that human papillomavirus (HPV) infection accounts for at least one-quarter of all HNSCCs. HPV+ and HPV- HNSCCs are studied as separate diseases as their prognosis, treatment, and molecular signatures are distinct. Five-year survival rates of HNSCC hover around 40-50%, and novel therapeutic targets and biomarkers are necessary to improve patient outcomes. Here, we investigate the DEK oncogene and its function in regulating HNSCC development and signaling. DEK is overexpressed in many cancer types, with roles in molecular processes such as transcription, DNA repair, and replication, as well as phenotypes such as apoptosis, senescence, and proliferation. DEK had never been previously studied in this tumor type; therefore, our studies began with clinical specimens to examine DEK expression patterns in primary HNSCC tissue. -

Memory Sequencing Reveals Heritable Single Cell Gene Expression Programs Associated with Distinct Cellular Behaviors
bioRxiv preprint doi: https://doi.org/10.1101/379016; this version posted July 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Title: Memory sequencing reveals heritable single cell gene expression programs associated with distinct cellular behaviors Authors: 1 2 1 1 2 Sydney M. Shaffer , Benjamin L. Emert , Ann E. Sizemore , Rohit Gupte , Eduardo Torre , 1,4,5,6 1,3 Danielle S. Bassett , Arjun Raj 1 D epartment of Bioengineering, School of Engineering and Applied Sciences, University of Pennsylvania 2 P erelman School of Medicine, University of Pennsylvania 3 D epartment of Genetics, Perelman School of Medicine, University of Pennsylvania 4 D epartment of Physics & Astronomy, School of Arts and Sciences, University of Pennsylvania 5 D epartment of Electrical & Systems Engineering, School of Engineering and Applied Sciences, University of Pennsylvania 6 D epartment of Neurology, Perelman School of Medicine, University of Pennsylvania Abstract: Non-genetic factors can cause individual cells to fluctuate substantially in gene expression levels over time. Yet it remains unclear whether these fluctuations can persist for much longer than the time for a single cell division. Current methods for measuring gene expression in single cells mostly rely on single time point measurements, making the time of a fluctuation of gene expression or cellular memory difficult to measure. Here, we report a method combining Luria and Delbrück’s fluctuation analysis with population-based RNA sequencing (MemorySeq) for identifying genes transcriptome-wide whose fluctuations persist for several cell divisions.