Hepatic Peroxisome and Its Implications on Cardiac Function, Inflammation and Aging
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

What Are Their Roles in Mitochondrial Protein Synthesis?
Characterisation of human mtRF1 and C12orf65: What are their roles in mitochondrial protein synthesis? Aleksandra Pajak M.Res Thesis submitted to Newcastle University in candidature for the degree of Doctor of Philosophy Newcastle University Faculty of Medical Sciences Institute for Ageing and Health Mitochondrial Research Group January 2013 Abstract Mitochondria have their own protein synthesis machinery that synthesises the oxidative phosphorylation components encoded by their mtDNA. This translation process consists of four main phases: initiation, elongation, termination and ribosome recycling. Termination and its control have been the least investigated. Recently, however, the termination factor, mtRF1a, has been characterised as sufficient to release all the nascent proteins from the mitoribosome. Furthermore, bioinformatics has identified three additional members of this mitochondrial release factor family namely, mtRF1, C12orf65 and ICT1. The latter is now known to be incorporated into the mitoribosome but its exact function remains unclear. My project has therefore focussed on characterising the remaining two factors; mtRF1 and C12orf65, and investigating their possible involvement in mitochondrial protein synthesis. It has been demonstrated that protein synthesis is not perfect and bacterial ribosomes not infrequently stall during translation. This can result from limiting amounts of charged tRNAs, stable secondary structures, or truncated/degraded transcripts. Ribosome stalling has been shown to cause growth arrest. In order to prevent that and maintain high efficiency of mitochondrial protein synthesis such stalled complexes need to be rapidly recycled. Bacteria have developed at least three distinct mechanisms by which ribosomes can be rescued. Contrastingly, despite the presence of truncated mRNAs in mitochondria, no such quality control mechanisms have been identified in these organelles. -

Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation
From THE DEPARTMENT OF MICROBIOLOGY TUMOR AND CELL BIOLOGY (MTC) Karolinska Institutet, Stockholm, Sweden ROLE OF MITOCHONDRIAL RIBOSOMAL PROTEIN S18-2 IN CANCEROGENESIS AND IN REGULATION OF STEMNESS AND DIFFERENTIATION Muhammad Mushtaq Stockholm 2017 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by E-Print AB 2017 © Muhammad Mushtaq, 2017 ISBN 978-91-7676-697-2 Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation THESIS FOR DOCTORAL DEGREE (Ph.D.) By Muhammad Mushtaq Principal Supervisor: Faculty Opponent: Associate Professor Elena Kashuba Professor Pramod Kumar Srivastava Karolinska Institutet University of Connecticut Department of Microbiology Tumor and Cell Center for Immunotherapy of Cancer and Biology (MTC) Infectious Diseases Co-supervisor(s): Examination Board: Professor Sonia Lain Professor Ola Söderberg Karolinska Institutet Uppsala University Department of Microbiology Tumor and Cell Department of Immunology, Genetics and Biology (MTC) Pathology (IGP) Professor George Klein Professor Boris Zhivotovsky Karolinska Institutet Karolinska Institutet Department of Microbiology Tumor and Cell Institute of Environmental Medicine (IMM) Biology (MTC) Professor Lars-Gunnar Larsson Karolinska Institutet Department of Microbiology Tumor and Cell Biology (MTC) Dedicated to my parents ABSTRACT Mitochondria carry their own ribosomes (mitoribosomes) for the translation of mRNA encoded by mitochondrial DNA. The architecture of mitoribosomes is mainly composed of mitochondrial ribosomal proteins (MRPs), which are encoded by nuclear genomic DNA. Emerging experimental evidences reveal that several MRPs are multifunctional and they exhibit important extra-mitochondrial functions, such as involvement in apoptosis, protein biosynthesis and signal transduction. Dysregulations of the MRPs are associated with severe pathological conditions, including cancer. -

Integrated Bioinformatic Analysis of RNA Binding Proteins in Hepatocellular Carcinoma
www.aging-us.com AGING 2021, Vol. 13, No. 2 Research Paper Integrated bioinformatic analysis of RNA binding proteins in hepatocellular carcinoma Ling Wang1,*, Zhen Zhang2,3,*, Yuan Li1, Yanyan Wan1, Baocai Xing1,& 1Key Laboratory of Carcinogenesis and Translational Research, Ministry of Education, Hepatopancreatobiliary Surgery Department I, Peking University Cancer Hospital and Institute, Beijing 100142, China 2Department of Gastroenterological Surgery, Peking University People’s Hospital, Beijing 100044, China 3Laboratory of Surgical Oncology, Beijing Key Laboratory of Colorectal Cancer Diagnosis and Treatment Research, Peking University People’s Hospital, Beijing 100044, China *Equal contribution Correspondence to: Baocai Xing; email: [email protected] Keywords: RNA binding protein, hepatocellular carcinoma, biomarker, transcriptomics, proteomics Received: June 8, 2020 Accepted: November 3, 2020 Published: December 19, 2020 Copyright: © 2020 Wang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT RNA binding proteins (RBPs) are aberrantly expressed in a tissue-specific manner across many tumors. These proteins, which play a vital role in post-transcriptional gene regulation, are involved in RNA splicing, maturation, transport, stability, degradation, and translation. We set out to establish an accurate risk score model based on RBPs to estimate prognosis in hepatocellular carcinoma (HCC). RNA-sequencing data, proteomic data and corresponding clinical information were acquired from the Cancer Genome Atlas database and the Clinical Proteomic Tumor Analysis Consortium database respectively. We identified 406 differentially expressed RBPs between HCC tumor and normal tissues at the transcriptional and protein level. -
![MRPS11 Mouse Monoclonal Antibody [Clone ID: OTI3D2] Product Data](https://docslib.b-cdn.net/cover/0941/mrps11-mouse-monoclonal-antibody-clone-id-oti3d2-product-data-210941.webp)
MRPS11 Mouse Monoclonal Antibody [Clone ID: OTI3D2] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for CF802839 MRPS11 Mouse Monoclonal Antibody [Clone ID: OTI3D2] Product data: Product Type: Primary Antibodies Clone Name: OTI3D2 Applications: IHC, WB Recommended Dilution: WB 1:2000 Reactivity: Human Host: Mouse Isotype: IgG1 Clonality: Monoclonal Immunogen: Full length human recombinant protein of human MRPS11 (NP_073750) produced in E.coli. Formulation: Lyophilized powder (original buffer 1X PBS, pH 7.3, 8% trehalose) Reconstitution Method: For reconstitution, we recommend adding 100uL distilled water to a final antibody concentration of about 1 mg/mL. To use this carrier-free antibody for conjugation experiment, we strongly recommend performing another round of desalting process. (OriGene recommends Zeba Spin Desalting Columns, 7KMWCO from Thermo Scientific) Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 20.4 kDa Gene Name: Homo sapiens mitochondrial ribosomal protein S11 (MRPS11), transcript variant 1, mRNA; nuclear gene for mitochondrial product. Database Link: NP_073750 Entrez Gene 64963 Human P82912 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 MRPS11 Mouse Monoclonal Antibody [Clone ID: OTI3D2] – CF802839 Background: Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. -

Analysis of the Relationship Between Ribosomal Protein and SSU Processome Assembly in Saccharomyces Cerevisiae
Analysis of the relationship between ribosomal protein and SSU processome assembly in Saccharomyces cerevisiae Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der naturwissenschaftlichen Fakultät III – Biologie und vorklinische Medizin - der Universität Regensburg vorgelegt von Steffen Jakob aus Wolfen Januar 2010 Promotionsgesuch eingereicht am: 13. Januar 2010 Die Arbeit wurde angeleitet von: Prof. Dr. Herbert Tschochner Prüfungsausschuss: Vorsitzender: Prof. Dr. Armin Kurtz 1. Prüfer: Prof. Dr. Herbert Tschochner 2. Prüfer: Prof. Dr. Rainer Deutzmann 3. Prüfer: Prof. Dr. Wolfgang Seufert Tag der mündlichen Prüfung: 24. März 2010 Die vorliegende Arbeit wurde in der Zeit von April 2006 bis Januar 2010 am Lehrstuhl Biochemie III des Institutes für Biochemie, Genetik und Mikrobiologie der Naturwissenschaftlichen Fakultät III der Universität zu Regensburg unter Anleitung von Dr. Philipp Milkereit im Labor von Prof. Dr. Herbert Tschochner angefertigt. Ich erkläre hiermit, dass ich diese Arbeit selbst verfasst und keine anderen als die angegebenen Quellen und Hilfsmittel verwendet habe. Diese Arbeit war bisher noch nicht Bestandteil eines Prüfungsverfahrens. Andere Promotionsversuche wurden nicht unternommen. Regensburg, den 13. Januar 2010 Steffen Jakob Table of Contents Table of Contents 1 SUMMARY ...................................................................................................... 1 2 INTRODUCTION ............................................................................................ -

MRPS11 Antibody (Center) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP17559C
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 MRPS11 Antibody (Center) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP17559C Specification MRPS11 Antibody (Center) - Product Information Application WB,E Primary Accession P82912 Other Accession NP_073750.2, NP_789775.1 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 20616 Antigen Region 42-69 MRPS11 Antibody (Center) - Additional Information MRPS11 Antibody (Center) (Cat. #AP17559c) western blot analysis in NCI-H460 cell line Gene ID 64963 lysates (35ug/lane).This demonstrates the Other Names MRPS11 antibody detected the MRPS11 28S ribosomal protein S11, mitochondrial, protein (arrow). MRP-S11, S11mt, Cervical cancer proto-oncogene 2 protein, HCC-2, MRPS11, RPMS11 MRPS11 Antibody (Center) - Background Target/Specificity Mammalian mitochondrial ribosomal proteins This MRPS11 antibody is generated from are encoded by rabbits immunized with a KLH conjugated nuclear genes and help in protein synthesis synthetic peptide between 42-69 amino within the acids from the Central region of human mitochondrion. Mitochondrial ribosomes MRPS11. (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. Dilution They have an estimated WB~~1:1000 75% protein to rRNA composition compared to prokaryotic ribosomes, Format where this ratio is reversed. Another difference Purified polyclonal antibody supplied in PBS between mammalian with 0.09% (W/V) sodium azide. This mitoribosomes and prokaryotic ribosomes is antibody is purified through a protein A that the latter contain column, followed by peptide affinity a 5S rRNA. Among different species, the purification. proteins comprising the mitoribosome differ greatly in sequence, and Storage sometimes in Maintain refrigerated at 2-8°C for up to 2 biochemical properties, which prevents easy weeks. -
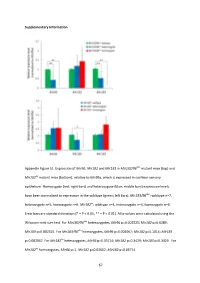
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

Predicting Clinical Response to Treatment with a Soluble Tnf-Antagonist Or Tnf, Or a Tnf Receptor Agonist
(19) TZZ _ __T (11) EP 2 192 197 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 02.06.2010 Bulletin 2010/22 C12Q 1/68 (2006.01) (21) Application number: 08170119.5 (22) Date of filing: 27.11.2008 (84) Designated Contracting States: (72) Inventor: The designation of the inventor has not AT BE BG CH CY CZ DE DK EE ES FI FR GB GR yet been filed HR HU IE IS IT LI LT LU LV MC MT NL NO PL PT RO SE SI SK TR (74) Representative: Habets, Winand Designated Extension States: Life Science Patents AL BA MK RS PO Box 5096 6130 PB Sittard (NL) (71) Applicant: Vereniging voor Christelijk Hoger Onderwijs, Wetenschappelijk Onderzoek en Patiëntenzorg 1081 HV Amsterdam (NL) (54) Predicting clinical response to treatment with a soluble tnf-antagonist or tnf, or a tnf receptor agonist (57) The invention relates to methods for predicting a clinical response to a therapy with a soluble TNF antagonist, TNF or a TNF receptor agonist and a kit for use in said methods. EP 2 192 197 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 192 197 A1 Description [0001] The invention relates to methods for predicting a clinical response to a treatment with a soluble TNF antagonist, with TNF or a TNF receptor agonist using expression levels of genes of the Type I INF pathway and a kit for use in said 5 methods. In another aspect, the invention relates to a method for evaluating a pharmacological effect of a treatment with a soluble TNF antagonist, TNF or a TNF receptor agonist. -

Rrna Types in Zebrafish Development
UvA-DARE (Digital Academic Repository) Distinct maternal and somatic rRNA types in zebrafish development Locati, M. Publication date 2017 Document Version Final published version License Other Link to publication Citation for published version (APA): Locati, M. (2017). Distinct maternal and somatic rRNA types in zebrafish development. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:04 Oct 2021 DISTINCT MATERNAL AND SOMATIC rRNA TYPES IN ZEBRAFISH DEVELOPMENT MAURO LOCATI DISTINCT MATERNAL AND SOMATIC rRNA TYPES IN ZEBRAFISH DEVELOPMENT MAURO LOCATI Distinct Maternal and Somatic rRNA Types in Zebrafish Development ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad van doctor aan de Universiteit van Amsterdam op gezag van de Rector Magnificus prof. dr. ir. K.I.J. Maex ten overstaan van een door het College voor Promoties ingestelde commissie, in het openbaar te verdedigen in de Aula der Universiteit op woensdag 6 december 2017, te 13:00 uur door Mauro Locati geboren te Palermo, Italië Promotiecommissie: Promotor: Prof.