Math Modeling for Undergraduates
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Chapter 5 Dimensional Analysis and Similarity
Chapter 5 Dimensional Analysis and Similarity Motivation. In this chapter we discuss the planning, presentation, and interpretation of experimental data. We shall try to convince you that such data are best presented in dimensionless form. Experiments which might result in tables of output, or even mul- tiple volumes of tables, might be reduced to a single set of curves—or even a single curve—when suitably nondimensionalized. The technique for doing this is dimensional analysis. Chapter 3 presented gross control-volume balances of mass, momentum, and en- ergy which led to estimates of global parameters: mass flow, force, torque, total heat transfer. Chapter 4 presented infinitesimal balances which led to the basic partial dif- ferential equations of fluid flow and some particular solutions. These two chapters cov- ered analytical techniques, which are limited to fairly simple geometries and well- defined boundary conditions. Probably one-third of fluid-flow problems can be attacked in this analytical or theoretical manner. The other two-thirds of all fluid problems are too complex, both geometrically and physically, to be solved analytically. They must be tested by experiment. Their behav- ior is reported as experimental data. Such data are much more useful if they are ex- pressed in compact, economic form. Graphs are especially useful, since tabulated data cannot be absorbed, nor can the trends and rates of change be observed, by most en- gineering eyes. These are the motivations for dimensional analysis. The technique is traditional in fluid mechanics and is useful in all engineering and physical sciences, with notable uses also seen in the biological and social sciences. -
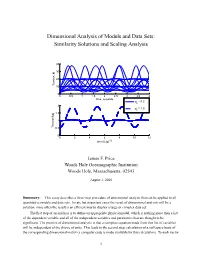
Dimensional Analysis of Models and Data Sets: Similarity Solutions and Scaling Analysis
Dimensional Analysis of Models and Data Sets: Similarity Solutions and Scaling Analysis 40 30 20 Tension, N 10 0 0 0.5 1 1.5 2 2.5 3 3.5 4 time, seconds φ = 0.2 o 2 φ = 1.0 o 1.5 1 Tension/Mg 0.5 0 0 2 4 6 8 10 12 time/(L/g)1/2 James F. Price Woods Hole Oceanographic Institution Woods Hole, Massachusetts, 02543 August 3, 2006 Summary: This essay describes a three-step procedure of dimensional analysis that can be applied to all quantitative models and data sets. In rare but important cases the result of dimensional analysis will be a solution; more often the result is an efficient way to display a large or complex data set. The first step of an analysis is to define an appropriate physical model, which is nothing more than a list of the dependent variable and all of the independent variables and parameters that are thought to be significant. The premise of dimensional analysis is that a complete equation made from this list of variables will be independent of the choice of units. This leads to the second step, calculation of a null space basis of the corresponding dimensional matrix (a computer code is made available for this calculation). To each vector 1 of the null space basis there corresponds a nondimensional variable, the number of which is less than the number of dimensional variables. The nondimensional variables are themselves a basis set and in most cases their form is not determined by dimensional analysis alone. -

Parametric Study of Magnetic Pendulum
Parametric Study of Magnetic Pendulum Author: Peter DelCioppo Persistent link: http://hdl.handle.net/2345/564 This work is posted on eScholarship@BC, Boston College University Libraries. Boston College Electronic Thesis or Dissertation, 2007 Copyright is held by the author, with all rights reserved, unless otherwise noted. Parametric Study of Magnetic Pendulum Boston College College of Arts and Sciences Honors Program Senior Thesis Project By Peter DelCioppo Advisor: Dr. Andrzej Herczynski, Physics Department Submitted May 2007 Table of Contents I. Genesis of Project…………………………………………………………………...p. 2 II Survey of Existing Literature…………………………………………………...…p. 2 III. Experimental Design……………………………………………………………...p. 3 IV. Theoretical Analysis………………………………………………………………p. 6 V. Calibration of Experimental Apparatus………………………………………….p. 8 VI. Experimental Results…………………………………………………………....p. 12 VII. Summary………………………………………………………………………..p. 15 VIII. References……………………………………………………………………...p. 17 - 1 - I. Genesis of Project This thesis project has been selected as an extension of experiments and research performed as part of the Spring 2006 Introduction to Chaos & Nonlinear Dynamics course (PH545). The current work particularly relates to experimental work analyzing the dynamics of the double well chaotic pendulum and a final project surveying the magnetic pendulum. In the experimental work, a basic understanding of the chaotic pendulum was developed, while the final project included a survey of the basic components of a magnetic pendulum system with particular -

Dimensional Analysis and Modeling
cen72367_ch07.qxd 10/29/04 2:27 PM Page 269 CHAPTER DIMENSIONAL ANALYSIS 7 AND MODELING n this chapter, we first review the concepts of dimensions and units. We then review the fundamental principle of dimensional homogeneity, and OBJECTIVES Ishow how it is applied to equations in order to nondimensionalize them When you finish reading this chapter, you and to identify dimensionless groups. We discuss the concept of similarity should be able to between a model and a prototype. We also describe a powerful tool for engi- I Develop a better understanding neers and scientists called dimensional analysis, in which the combination of dimensions, units, and of dimensional variables, nondimensional variables, and dimensional con- dimensional homogeneity of equations stants into nondimensional parameters reduces the number of necessary I Understand the numerous independent parameters in a problem. We present a step-by-step method for benefits of dimensional analysis obtaining these nondimensional parameters, called the method of repeating I Know how to use the method of variables, which is based solely on the dimensions of the variables and con- repeating variables to identify stants. Finally, we apply this technique to several practical problems to illus- nondimensional parameters trate both its utility and its limitations. I Understand the concept of dynamic similarity and how to apply it to experimental modeling 269 cen72367_ch07.qxd 10/29/04 2:27 PM Page 270 270 FLUID MECHANICS Length 7–1 I DIMENSIONS AND UNITS 3.2 cm A dimension is a measure of a physical quantity (without numerical val- ues), while a unit is a way to assign a number to that dimension. -

Advanced Programming in Engineering Lecture Notes
Advanced Programming in Engineering lecture notes P.J. Visser, S. Luding, S. Srivastava March 13, 2011 Changes 13/3/2011 Added Section 7.4 to 7.7. Added Section 4.4 7/3/2011 Added chapter Chapter 7 23/2/2011 Corrected the equation for the average temperature in terms of micro- scopic quantities, Eq. 3.14. Corrected the equation for the speed distribution in 2D, Eq. 3.19. Added links to sources of `true' random numbers available through the web as footnotes in Section 5.1. Added examples of choices for parameters of the LCG and LFG in Section 5.1.1 and 5.1.2, respectively. Added the chi-square test and correlation functions as ways for testing random numbers in Section 5.1.4. Improved the derivation of the average squared distance of particles in a random walk being linear with time, in Section 5.2.1. Added a discussion of the meaning of the terms in the master equation, in Section 5.2.5. Small changes. 19/2/2011 Corrected the Maxwell-Boltzmann distribution for the speed, Eq. 3.19. 14/2/2011 1 2 Added Chapter 6. 6/2/2011 Verified Eq. 3.15 and added footnote discussing its meaning in case of periodic boundary conditions. Added formula for the radial distribution function in Section 3.4.2, Eq. 3.21. 31/1/2011 Added Chapter 5. 9/1/2010 Corrected labels on the axes in Fig. 3.10, changed U · " and r · σ to U=" and r/σ, respectively. Added truncated version of the Lennard-Jones potential in Section 3.3.1. -

Dimensional Analysis and Modeling
cen72367_ch07.qxd 10/29/04 2:27 PM Page 269 CHAPTER DIMENSIONAL ANALYSIS 7 AND MODELING n this chapter, we first review the concepts of dimensions and units. We then review the fundamental principle of dimensional homogeneity, and OBJECTIVES Ishow how it is applied to equations in order to nondimensionalize them When you finish reading this chapter, you and to identify dimensionless groups. We discuss the concept of similarity should be able to between a model and a prototype. We also describe a powerful tool for engi- ■ Develop a better understanding neers and scientists called dimensional analysis, in which the combination of dimensions, units, and of dimensional variables, nondimensional variables, and dimensional con- dimensional homogeneity of equations stants into nondimensional parameters reduces the number of necessary ■ Understand the numerous independent parameters in a problem. We present a step-by-step method for benefits of dimensional analysis obtaining these nondimensional parameters, called the method of repeating ■ Know how to use the method of variables, which is based solely on the dimensions of the variables and con- repeating variables to identify stants. Finally, we apply this technique to several practical problems to illus- nondimensional parameters trate both its utility and its limitations. ■ Understand the concept of dynamic similarity and how to apply it to experimental modeling 269 cen72367_ch07.qxd 10/29/04 2:27 PM Page 270 270 FLUID MECHANICS Length 7–1 ■ DIMENSIONS AND UNITS 3.2 cm A dimension is a measure of a physical quantity (without numerical val- ues), while a unit is a way to assign a number to that dimension. -

Aircraft Interior Noise Reduction by Alternate Resonance Tuning Progress Report for the Period Ending December, 1990 Final Progr
_," ;'if ,// ... ..<-. T:- "-"" AIRCRAFT INTERIOR NOISE REDUCTION BY ALTERNATE RESONANCE TUNING PROGRESS REPORT FOR THE PERIOD ENDING DECEMBER, 1990 FINAL PROGRESS REPORT NASA RESEARCH GRANT # NAG-I-722 Prepared for: STRUCTURAL ACOUSTICS BRANCH NASA LANGLEY RESEARCH CENTER Prepared by: Dr. James A. Gottwald Dr. Donald B. Bliss Department of Mechanical Engineering and Materials Science DUKE UNIVERSITY DURHAM, NC 27705 N <) L-/S ;:: "7 7 ..... !L -, r TbN L_io .... • k t,Jniv ) 2:_-, i- GSCL 2_7)tt Abstract Existing interior noise reduction techniques for aircraft fuselages perform reasonably well at higher frequencies, but are inadequate at lower frequencies, particularly with respect to the low blade passage harmonics with high forcing levels found in propeller aircraft. This research focuses on a noise control method which considers aircraft fuselages lined with panels alternately tuned to frequencies above and below the frequency that must be attenuated. Adjacent panels would oscillate at equal amplitude, to give equal source strength, but with opposite phase. Provided these adjacent panels are acoustically compact, the resulting cancellation causes the interior acoustic modes to become cutoff, and therefore be non-propagating and evanescent. This interior noise reduction method, called Alternate Resonance Tuning (ART), is described in this thesis both theoretically and experimentally. Problems presented herein deal with tuning single paneled wall structures for optimum noise reduction using the ART tuning methodology, and three theoretical problems are analyzed. The first analysis is a three dimensional, full acoustic solution for tuning a panel wall composed of repeating sections with four different panel tunings within that section, where the panels are modeled as idealized spring-mass-damper systems. -

Nondimensionalization, Scaling, and Units
Topic 1: Nondimensionalization, Scaling, and Units Course Notes, Math 558 Spring 2012 Barenblatt.book Holmes.new.book This section is a selection of material related to include Chapters 0–3 of [1], Chapters 1–3 of [2]. 1 Dimensional Analysis 1.1 Example 1. Throw a ball Consider the following example: We throw a ball directly upwards from the surface of the Earth. What is its maximum height? We need two physical laws to describe this system: 1. Newton’s Second Law. The acceleration of a body is directly proportional to its force and inversely proportional to its mass; moreover, the acceleration is parallel to the form. This is typically said as “Force equals mass times acceleration” or F = ma. 2. Newton’s law of universal gravitation. Every body in the universe attracts every other body with a gravitational force, which force is proportional to each of the masses of the objects and inversely proportional to the square of the distance between the objects. In equations, if x is the vector from mass 1 to mass 2, then the force on mass 1 due to mass 2 is Gm1m2x F12 = ; kxk3 and the force on mass 2 due to mass 1 is Gm1m2x F21 = −F12 = − : kxk3 Since this problem is one-dimensional, we can think of the force, acceleration, and position as scalars (dis- tance from the ground). So if we define mB as the mass of the ball, mE as the mass of the earth, then we have the differential equation for x(t), the height above the ground, as 2 d x GmBmE m = − ; (1) eq:d B dt2 d2 where d = R + x is the distanceeq:d between the center of the ball and the center of the earth (R is the radius of the earth). -

Introduction of Nonlinear Dynamics Into an Undergraduate Intermediate Dynamics Course
Introduction of Nonlinear Dynamics into a Undergraduate Intermediate Dynamics Course Bongsu Kang Department of Mechanical Engineering Indiana University – Purdue University Fort Wayne Abstract This paper presents a way to introduce nonlinear dynamics and numerical analysis tools to junior and senior mechanical engineering students through an intermediate dynamics course. The main purpose of introducing nonlinear dynamics into a undergraduate dynamics course is to increase the student awareness of the rich dynamic behavior of physical systems that mechanical engineers often encounter in real world applications, while a more rigorous and quantitative treatment of this subject is left to graduate-level dynamics and vibration courses. Employing relatively simple mechanical systems, well known nonlinear dynamics phenomena such as the jump phenomenon due to stiffness hardening/softening, self excitation/limit cycle, parametric resonance, irregular motion, and the basic concepts of stability are introduced. In addition, the students are introduced to essential analysis techniques such as phase diagrams, Poincaré maps, and nondimensionalization. Equations of motion governing such nonlinear systems are numerically integrated with the Matlab ODE solvers rather than analytically to ensure that the student’s comprehension of the subject is not hindered by mathematics which are beyond the typical undergraduate engineering curricula. Discussions of the subject in the course underline the qualitative behavior of nonlinear dynamic systems rather than quantitative analysis. Introduction Typical undergraduate mechanical engineering curricula layout a sequence of dynamics and vibration courses, e.g., dynamics (first dynamics course), kinematics and kinetics of machinery or similar courses, system dynamics, and relevant technical electives. The Bachelor of Science in Mechanical Engineering program at Indiana University – Purdue University Fort Wayne (IPFW) requires three core courses and offers two technical elective courses in the areas of dynamics and vibration. -

Course Notes, Math 558
Course Notes, Math 558 March 28, 2012 2 Contents 1 Nondimensionalization, Scaling, and Units 5 1.1 Dimensional Analysis . 5 1.1.1 Example 1. Throw a ball . 5 1.1.2 Example 2. Drag on a sphere . 7 1.1.3 Using dimensional analysis for scale models . 8 1.1.4 Buckingham Pi Theorem . 9 1.1.5 Example 3. Computing the yield of a nuclear device . 10 1.1.6 Example 4. Pythagoras’ Theorem . 11 1.1.7 Example 5. Diffusion equation . 11 1.2 Scaling and nondimensionalization . 12 1.2.1 Projectile problem revisited . 12 1.2.2 A different scaling for projectile problem . 14 1.2.3 Diffusion equation, revisited . 14 2 Regular Asymptotics 17 2.1 Motivation, definitions . 17 2.2 Asymptotics for polynomials . 18 2.2.1 Examples . 18 2.2.2 Systematic expansions for polynomials . 19 2.3 Asymptotics for Differential Equations . 22 2.3.1 Examples . 22 2.3.2 Regular asymptotic regime . 24 3 Random Walks, Diffusions, and SDE 27 3.1 Random Walks . 27 3.1.1 Unbiased random walk — combinatorial approach . 27 3.1.2 Unbiased random walk— asymptotic approach . 29 3.1.3 Biased random walk . 30 3.1.4 Another unbiased random walk . 31 3.1.5 Summary . 31 3.2 Solution of heat equation . 31 3.2.1 No drift term . 31 3.2.2 Drift term . 33 3.3 Stochastic differential equations . 33 3.3.1 Background on probability theory . 33 3.3.2 Stochastic differential equations . 34 3.3.3 Fokker-Planck equations . 35 3.4 Comments on units, scaling . -
Enhanced Closed Loop Performance Using Non-Dimensional Analysis
Rochester Institute of Technology RIT Scholar Works Theses 2004 Enhanced closed loop performance using non-dimensional analysis Michael H. Wilson Follow this and additional works at: https://scholarworks.rit.edu/theses Recommended Citation Wilson, Michael H., "Enhanced closed loop performance using non-dimensional analysis" (2004). Thesis. Rochester Institute of Technology. Accessed from This Thesis is brought to you for free and open access by RIT Scholar Works. It has been accepted for inclusion in Theses by an authorized administrator of RIT Scholar Works. For more information, please contact [email protected]. Enhanced Closed Loop Performance Using Non-Dimensional Analysis By Michael H. Wilson A Thesis Submitted in Partial Fulfillment of the Requirement for the MASTER OF SCIENCE IN MECHANICAL ENGINEERING Approved by: Dr. Agamemnon Crassidis (Thesis Advisor) Department of Mechanical Engineering Dr. Mark Kemski Department of Mechanical Engineering Dr. Josef Torok Department of Mechanical Engineering Dr. Edward C. Hensel Department Head of Mechanical Engineering DEPARTMENT OF MECHANICAL ENGINEERING ROCHESTER INSTITUTE OF TECHNOLOGY DECEMBER, 2004 Enhanced Closed-Loop Performance Using Non-Dimensional Analysis I, Michael H. Wilson, hereby grant permission for the Wallace Library of the Rochester Institute ofTechnology to reproduce my thesis in whole or in part. Any reproduction will not be for commercial use or profit. Signature of Author:. Michael H. Wi lson Date: JL Ii 3•Ie 1_ 11 ABSTRACT This paper investigates the benefits of using non-dimension analysis to develop a control law for a flexible electro-mechanical system. The system that is analyzed consists of a DC motor connected to a load inertia through a set of gears. -
Numerical Methods for the Unsteady Compressible Navier-Stokes Equations
Numerical Methods for the Unsteady Compressible Navier-Stokes Equations Philipp Birken Numerical Methods for the Unsteady Compressible Navier-Stokes Equations Dr. Philipp Birken Habilitationsschrift am Fachbereich Mathematik und Naturwissenschaften der Universit¨atKassel Gutachter: Prof. Dr. Andreas Meister Zweitgutachterin: Prof. Dr. Margot Gerritsen Drittgutachterin: Prof. Dr. Hester Bijl Probevorlesung: 31. 10. 2012 3 There are all kinds of interesting questions that come from a know- ledge of science, which only adds to the excitement and mystery and awe of a flower. It only adds. I don't un- derstand how it subtracts. Richard Feynman 6 Acknowledgements A habilitation thesis like this is not possible without help from many people. First of all, I'd like to mention the German Research Foundation (DFG), which has funded my research since 2006 as part of the collaborative research area SFB/TR TRR 30, project C2. This interdisciplinary project has provided me the necessary freedom, motivation and optimal funding to pursue the research documented in this book. It also allowed me to hire students to help me in my research and I'd like to thank Benjamin Badel, Maria Bauer, Veronika Diba, Marouen ben Said and Malin Trost, who did a lot of coding and testing and created a lot of figures. Furthermore, I thank the colleagues from the TRR 30 for a very successful research cooperation and in particular Kurt Steinhoff for starting the project. Then, there are my collaborators over the last years, in alphabetical order Hester Bijl, who thankfully also acted as reviewer, Jurjen Duintjer Tebbens, Gregor Gassner, Mark Haas, Volker Hannemann, Stefan Hartmann, Antony Jameson, Detlef Kuhl, Andreas Meis- ter, Claus-Dieter Munz, Karsten J.