The Parkinson Disease Gene SNCA
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Parkin Antibody (C-Term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP6402B
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Parkin Antibody (C-term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP6402B Specification Parkin Antibody (C-term) - Product Information Application IF, WB, IHC-P, FC,E Primary Accession O60260 Other Accession NP_004553 Reactivity Human, Mouse Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Antigen Region 387-417 Parkin Antibody (C-term) - Additional Information Gene ID 5071 Other Names E3 ubiquitin-protein ligase parkin, 632-, Confocal immunofluorescent analysis of Parkinson juvenile disease protein 2, Parkin Antibody (C-term)(Cat#AP6402b) with Parkinson disease protein 2, PARK2, PRKN NCI-H460 cell followed by Alexa Fluor 488-conjugated goat anti-rabbit lgG Target/Specificity (green).Actin filaments have been labeled This Parkin antibody is generated from with Alexa Fluor 555 phalloidin (red).DAPI rabbits immunized with a KLH conjugated was used to stain the cell nuclear (blue). synthetic peptide between 387-417 amino acids from the C-terminal region of human Parkin. Dilution IF~~1:10~50 WB~~1:1000 IHC-P~~1:50~100 FC~~1:10~50 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C Park2 Antibody (C-term) (Cat. #AP6402b) in small aliquots to prevent freeze-thaw western blot analysis in K562 cell line lysates cycles. (35ug/lane).This demonstrates the Park2 antibody detected the Park2 protein (arrow). -

Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen
CLINICAL RESEARCH www.jasn.org Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen Rany M. Salem ,1 Jennifer N. Todd,2,3,4 Niina Sandholm ,5,6,7 Joanne B. Cole ,2,3,4 Wei-Min Chen,8 Darrell Andrews,9 Marcus G. Pezzolesi,10 Paul M. McKeigue,11 Linda T. Hiraki,12 Chengxiang Qiu,13 Viji Nair,14 Chen Di Liao,12 Jing Jing Cao,12 Erkka Valo ,5,6,7 Suna Onengut-Gumuscu,8 Adam M. Smiles,15 Stuart J. McGurnaghan,16 Jani K. Haukka,5,6,7 Valma Harjutsalo,5,6,7,17 Eoin P. Brennan,9 Natalie van Zuydam,18,19 Emma Ahlqvist,20 Ross Doyle,9 Tarunveer S. Ahluwalia ,21 Maria Lajer,21 Maria F. Hughes,9 Jihwan Park,13 Jan Skupien,15 Athina Spiliopoulou,11 Andrew Liu,22 Rajasree Menon,14,23 Carine M. Boustany-Kari,24 Hyun M. Kang,23,25 Robert G. Nelson,26 Ronald Klein,27 Barbara E. Klein,27 Kristine E. Lee ,27 Xiaoyu Gao,28 Michael Mauer,29 Silvia Maestroni,30 Maria Luiza Caramori,29 Ian H. de Boer ,31 Rachel G. Miller,32 Jingchuan Guo ,32 Andrew P. Boright,12 David Tregouet,33,34 Beata Gyorgy,33,34 Janet K. Snell-Bergeon,35 David M. Maahs,36 Shelley B. Bull ,37 Angelo J. Canty,38 Colin N.A. Palmer,39 Lars Stechemesser,40 Bernhard Paulweber,40 Raimund Weitgasser,40,41 Jelizaveta Sokolovska,42 Vita Rovıte,43 Valdis Pırags, 42,44 Edita Prakapiene,45 Lina Radzeviciene,46 Rasa Verkauskiene,46 Nicolae Mircea Panduru,6,47 Leif C. -

Reading Through the Building Blocks of the Genome: Exonic Variation in PD
UNIVERSITY OF CATANIA INTERNATIONAL PhD PROGRAM IN NEUROSCIENCE XXIX Cycle VALENTINA LA COGNATA READING THROUGH THE BUILDING BLOCKS OF THE GENOME: EXONIC VARIATION IN PARKINSON'S DISEASE PhD Thesis Coordinator: Supervisors: Prof. Salvatore Salomone Prof. Velia D’Agata Dr. Sebastiano Cavallaro January 2017 Copyright © V. La Cognata, 2017 All rights reserved. No part of this book may be reproduced, stored in a retrieval system or transmitted in any form or by any means, without prior permission of the author. The copyright of the published papers remains with the publishers. SUMMARY Abstract ...................................................................................................... 5 Chapter 1 .................................................................................................... 7 PARKINSON’S DISEASE ....................................................................................................... 9 HAS EXONIC VARIATION A ROLE IN PD? ............................................................................12 AIMS OF THE PhD WORK ...................................................................................................13 Chapter 2 .................................................................................................. 15 ABSTRACT ........................................................................................................................17 INTRODUCTION ................................................................................................................18 CNVs: A PREVALENT -

Subgroup-Specific Structural Variation Across 1,000 Medulloblastoma Genomes
ARTICLE doi:10.1038/nature11327 Subgroup-specific structural variation across 1,000 medulloblastoma genomes A list of authors and their affiliations appears at the end of the paper Medulloblastoma, the most common malignant paediatric brain tumour, is currently treated with nonspecific cytotoxic therapies including surgery, whole-brain radiation, and aggressive chemotherapy. As medulloblastoma exhibits marked intertumoural heterogeneity, with at least four distinct molecular variants, previous attempts to identify targets for therapy have been underpowered because of small samples sizes. Here we report somatic copy number aberrations (SCNAs) in 1,087 unique medulloblastomas. SCNAs are common in medulloblastoma, and are predominantly subgroup-enriched. The most common region of focal copy number gain is a tandem duplication of SNCAIP, a gene associated with Parkinson’s disease, which is exquisitely restricted to Group 4a. Recurrent translocations of PVT1, including PVT1-MYC and PVT1-NDRG1, that arise through chromothripsis are restricted to Group 3. Numerous targetable SCNAs, including recurrent events targeting TGF-b signalling in Group 3, and NF-kB signalling in Group 4, suggest future avenues for rational, targeted therapy. Brain tumours are the most common cause of childhood oncological Twenty-eight regions of recurrent high-level amplification (copy death, and medulloblastoma is the most common malignant paediatric number $ 5) were identified (Fig. 1d and Supplementary Table 7). brain tumour. Current medulloblastoma therapy including surgical The most prevalent amplifications affected members of the MYC resection, whole-brain and spinal cord radiation, and aggressive family with MYCN predominantly amplified in SHH and Group 4, chemotherapy supplemented by bone marrow transplant yields five- MYC in Group 3, and MYCL1 in SHH medulloblastomas. -
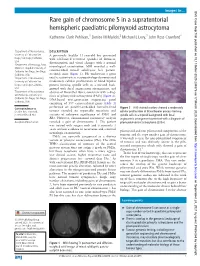
Rare Gain of Chromosome 5 in a Supratentorial Hemispheric
Images in… BMJ Case Rep: first published as 10.1136/bcr-2020-234878 on 17 March 2020. Downloaded from Rare gain of chromosome 5 in a supratentorial hemispheric paediatric pilomyxoid astrocytoma Katherine Clark Pehlivan,1 Denise M Malicki,2 Michael L Levy,3 John Ross Crawford4 1Department of Neurosciences, DESCRIPTION University of California San A previously healthy 11- year- old boy presented Diego, San Diego, California, with self- limited recurrent episodes of dizziness, USA disorientation and visual changes with a normal 2Department of Pathology, Rady neurological examination. MRI revealed a well- Children’s Hospital University of California San Diego, San Diego, circumscribed mixed solid/cystic left parieto- California, USA occipital mass (figure 1). He underwent a gross 3Department of Neurosurgery, total resection where neuropathology demonstrated University of California San moderately cellular proliferation of bland bipolar Diego, San Diego, California, process- forming spindle cells in a myxoid back- USA ground with focal angiocentric arrangement, and 4 Department of Neurosciences absence of Rosenthal fibres, consistent with a diag- and Pediatrics, University of nosis of pilomyxoid astrocytoma (PMA) (figure 2). California San Diego, San Diego, DNA- based next- generation sequencing panel California, USA consisting of 397 cancer- related genes (table 1) performed on paraffin- embedded formalin- fixed Correspondence to Figure 2 H&E- stained sections showed a moderately Dr John Ross Crawford; tumour revealed no reportable mutations and cellular proliferation of bland bipolar process- forming jrcrawford@ ucsd. edu variants of unknown significance of PMS1 and spindle cells in a myxoid background with focal RB1. However, chromosomal microarray analysis angiocentric arrangement consistent with a diagnosis of Accepted 4 March 2020 revealed a gain of chromosome 5. -

Identification of Significant Genes with Poor Prognosis in Ovarian Cancer Via Bioinformatical Analysis
Feng et al. Journal of Ovarian Research (2019) 12:35 https://doi.org/10.1186/s13048-019-0508-2 RESEARCH Open Access Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis Hao Feng1†, Zhong-Yi Gu2†, Qin Li2, Qiong-Hua Liu3, Xiao-Yu Yang4* and Jun-Jie Zhang2* Abstract Ovarian cancer (OC) is the highest frequent malignant gynecologic tumor with very complicated pathogenesis. The purpose of the present academic work was to identify significant genes with poor outcome and their underlying mechanisms. Gene expression profiles of GSE36668, GSE14407 and GSE18520 were available from GEO database. There are 69 OC tissues and 26 normal tissues in the three profile datasets. Differentially expressed genes (DEGs) between OC tissues and normal ovarian (OV) tissues were picked out by GEO2R tool and Venn diagram software. Next, we made use of the Database for Annotation, Visualization and Integrated Discovery (DAVID) to analyze Kyoto Encyclopedia of Gene and Genome (KEGG) pathway and gene ontology (GO). Then protein-protein interaction (PPI) of these DEGs was visualized by Cytoscape with Search Tool for the Retrieval of Interacting Genes (STRING). There were total of 216 consistently expressed genes in the three datasets, including 110 up-regulated genes enriched in cell division, sister chromatid cohesion, mitotic nuclear division, regulation of cell cycle, protein localization to kinetochore, cell proliferation and Cell cycle, progesterone-mediated oocyte maturation and p53 signaling pathway, while 106 down-regulated genes enriched in palate development, blood coagulation, positive regulation of transcription from RNA polymerase II promoter, axonogenesis, receptor internalization, negative regulation of transcription from RNA polymerase II promoter and no significant signaling pathways. -

Parkinson's Disease
Parkinson´s Disease Parkinson's disease (PD) is a chronic, progressive neurodegenerative disorder of the central nervous system characterized by tremor, muscle rigidity, postural abnormalities and slowed movement (bradykinesia), with dementia manifesting in 20 % of the afflicted. Onset before age 20 is categorized as juvenile-onset, before age 50 as early-onset and after age 50 as late-onset PD. After Alzheimer's disease, PD is the second most common neurodegenerative disorder. Symptoms occur from decreased stimulation of the motor cortex by the basal ganglia, resulting from insufficient formation of dopamine produced in the dopaminergic neurons in the midbrain (especially in the substantia nigra). Mutations in SNCA, UCHL1, PARK8/LRRK2 and PARK3 result in autosomal dominant PD, while mutations in PARK2, PARK7, and PINK1 result in autosomal recessive PD. NR4A2 and SNCAIP have been identified as susceptibility for PD. Alpha-synuclein (SNCA) is an acidic neuronal protein. Parkin coregulated gene protein (PACRG) is a gene Accumulation of insoluble fibrils of aggregated SNCA located close to parkin and thought to be co-transcribed protein is found in the central pathologic feature of PD, the with parkin. The PACRG protein is also a component of Lewy bodies. Therefore, PD is generally considered a Lewy bodies in PD patients. synucleinopathy. DJ-1/PARK7 participates in protection of neuronal cells from oxidative stress and mutation may lead to loss of this protection causing neuronal degradation. The roles of oxidative stress and mitochondrial dysfunction are under investigation as contributing factors to onset of PD. IHC staining using SNCA mouse mono- clonal antibody TA506533 of paraffin- embedded Human embryonic cerebellum PTEN-induced putative kinase 1 (PINK1) is a Ubiquitin carboxy-terminal hydrolase L1 (UCHL1) is a mitochondrial serine/threonine-protein kinase thought deubiquitinating enzyme. -

Entrez ID Gene Name Fold Change Q-Value Description
Entrez ID gene name fold change q-value description 4283 CXCL9 -7.25 5.28E-05 chemokine (C-X-C motif) ligand 9 3627 CXCL10 -6.88 6.58E-05 chemokine (C-X-C motif) ligand 10 6373 CXCL11 -5.65 3.69E-04 chemokine (C-X-C motif) ligand 11 405753 DUOXA2 -3.97 3.05E-06 dual oxidase maturation factor 2 4843 NOS2 -3.62 5.43E-03 nitric oxide synthase 2, inducible 50506 DUOX2 -3.24 5.01E-06 dual oxidase 2 6355 CCL8 -3.07 3.67E-03 chemokine (C-C motif) ligand 8 10964 IFI44L -3.06 4.43E-04 interferon-induced protein 44-like 115362 GBP5 -2.94 6.83E-04 guanylate binding protein 5 3620 IDO1 -2.91 5.65E-06 indoleamine 2,3-dioxygenase 1 8519 IFITM1 -2.67 5.65E-06 interferon induced transmembrane protein 1 3433 IFIT2 -2.61 2.28E-03 interferon-induced protein with tetratricopeptide repeats 2 54898 ELOVL2 -2.61 4.38E-07 ELOVL fatty acid elongase 2 2892 GRIA3 -2.60 3.06E-05 glutamate receptor, ionotropic, AMPA 3 6376 CX3CL1 -2.57 4.43E-04 chemokine (C-X3-C motif) ligand 1 7098 TLR3 -2.55 5.76E-06 toll-like receptor 3 79689 STEAP4 -2.50 8.35E-05 STEAP family member 4 3434 IFIT1 -2.48 2.64E-03 interferon-induced protein with tetratricopeptide repeats 1 4321 MMP12 -2.45 2.30E-04 matrix metallopeptidase 12 (macrophage elastase) 10826 FAXDC2 -2.42 5.01E-06 fatty acid hydroxylase domain containing 2 8626 TP63 -2.41 2.02E-05 tumor protein p63 64577 ALDH8A1 -2.41 6.05E-06 aldehyde dehydrogenase 8 family, member A1 8740 TNFSF14 -2.40 6.35E-05 tumor necrosis factor (ligand) superfamily, member 14 10417 SPON2 -2.39 2.46E-06 spondin 2, extracellular matrix protein 3437 -

Robles JTO Supplemental Digital Content 1
Supplementary Materials An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA and DNA Methylation Biomarkers Ana I. Robles1, Eri Arai2, Ewy A. Mathé1, Hirokazu Okayama1, Aaron Schetter1, Derek Brown1, David Petersen3, Elise D. Bowman1, Rintaro Noro1, Judith A. Welsh1, Daniel C. Edelman3, Holly S. Stevenson3, Yonghong Wang3, Naoto Tsuchiya4, Takashi Kohno4, Vidar Skaug5, Steen Mollerup5, Aage Haugen5, Paul S. Meltzer3, Jun Yokota6, Yae Kanai2 and Curtis C. Harris1 Affiliations: 1Laboratory of Human Carcinogenesis, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 2Division of Molecular Pathology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 3Genetics Branch, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 4Division of Genome Biology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 5Department of Chemical and Biological Working Environment, National Institute of Occupational Health, NO-0033 Oslo, Norway. 6Genomics and Epigenomics of Cancer Prediction Program, Institute of Predictive and Personalized Medicine of Cancer (IMPPC), 08916 Badalona (Barcelona), Spain. List of Supplementary Materials Supplementary Materials and Methods Fig. S1. Hierarchical clustering of based on CpG sites differentially-methylated in Stage I ADC compared to non-tumor adjacent tissues. Fig. S2. Confirmatory pyrosequencing analysis of DNA methylation at the HOXA9 locus in Stage I ADC from a subset of the NCI microarray cohort. 1 Fig. S3. Methylation Beta-values for HOXA9 probe cg26521404 in Stage I ADC samples from Japan. Fig. S4. Kaplan-Meier analysis of HOXA9 promoter methylation in a published cohort of Stage I lung ADC (J Clin Oncol 2013;31(32):4140-7). Fig. S5. Kaplan-Meier analysis of a combined prognostic biomarker in Stage I lung ADC. -

(SNCAIP) (NM 005460) Human 3' UTR Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC207980 Synphilin 1 (SNCAIP) (NM_005460) Human 3' UTR Clone Product data: Product Type: 3' UTR Clones Product Name: Synphilin 1 (SNCAIP) (NM_005460) Human 3' UTR Clone Vector: pMirTarget (PS100062) Symbol: SNCAIP Synonyms: Sph1; SYPH1 ACCN: NM_005460 Insert Size: 753 bp Insert Sequence: >SC207980 3’UTR clone of NM_005460 The sequence shown below is from the reference sequence of NM_005460. The complete sequence of this clone may contain minor differences, such as SNPs. Blue=Stop Codon Red=Cloning site GGCAAGTTGGACGCCCGCAAGATCCGCGAGATTCTCATTAAGGCCAAGAAGGGCGGAAAGATCGCCGTG TAACAATTGGCAGAGCTCAGAATTCAAGCGATCGCC GCTAGCAAAGGAAAGAATAAGGCAGCATAATGACATCAATAGAAAAATGAAGAAATCCTACAGCATAAA GCACATTGCTGAGCCAGAGTCAAAAGAACTCTTCTTGTAAATCACTTTTTAAATTTTCTCTCACTGATG CCCTTTGGAAATTATTGGAAATTTCTGGACTATCCTCTTTGGAAAGAGAACCATGAAAACAATGCCTCA CCAGCAGAAGAACAGAATATCAGGATGCCTTAAATTTATAGTAGTAGACTGTAAAAGATTCATTTTGGG GTGATATCTGTATATATAACTTGTTTTTTTAAAAGATGCCGTTTAAAAGCATGATTGGGAAAATGTATG TTTTTTAAGAGTAGATTGATTCACCCTACCCACAGGACATTCACCAAGCCACTGATACCATTTTATATT TCATCAATTGCATGAGTATTTGCTAATGTTGATTGAACCTCCCTTTCCCCATAATGTGGGCAGATTTGG CTCAGCTCCTTCATGAGATCAGGTCAGTGGTATTGTTTCTGTCAAGAGTGTTTTTTCTGTCATTTCTAC TTTTTGTATAAAGGAAATAAAACAATGTTAACAGCCACCTATAAGCTTGGGGCACCTACTTTCTAACAA ATCTGTGATGTTCTGATTTCTAAGGGACTTTGCAAGTATTTTGATTGCTGATATTTGATTTCAGGAACA AACTGCTCAGTTTAGCAGTTTTCCCATAGGCGTCAAATAAAACATTCTATATTTCATTAAGTA -

Description: Uniprot:O60260 Alternative Names: Specificity
TA6500 Parkin Antibody Order 021-34695924 [email protected] Support 400-6123-828 50ul [email protected] 100 uL √ √ Web www.ab-mart.com.cn Description: Functions within a multiprotein E3 ubiquitin ligase complex, catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins, such as BCL2, SYT11, CCNE1, GPR37, RHOT1/MIRO1, MFN1, MFN2, STUB1, SNCAIP, SEPTIN5, TOMM20, USP30, ZNF746 and AIMP2. Mediates monoubiquitination as well as 'Lys-6', 'Lys-11', 'Lys-48'-linked and 'Lys-63'-linked polyubiquitination of substrates depending on the context. Participates in the removal and/or detoxification of abnormally folded or damaged protein by mediating 'Lys-63'-linked polyubiquitination of misfolded proteins such as PARK7: 'Lys-63'-linked polyubiquitinated misfolded proteins are then recognized by HDAC6, leading to their recruitment to aggresomes, followed by degradation. Mediates 'Lys-63'-linked polyubiquitination of a 22 kDa O-linked glycosylated isoform of SNCAIP, possibly playing a role in Lewy-body formation. Mediates monoubiquitination of BCL2, thereby acting as a positive regulator of autophagy. Promotes the autophagic degradation of dysfunctional depolarized mitochondria (mitophagy) by promoting the ubiquitination of mitochondrial proteins such as TOMM20, RHOT1/MIRO1 and USP30. Preferentially assembles 'Lys-6'-, 'Lys-11'- and 'Lys-63'-linked polyubiquitin chains following mitochondrial damage, leading to mitophagy. Mediates 'Lys-48'-linked polyubiquitination of ZNF746, followed by degradation of ZNF746 by the proteasome; possibly playing a role in the regulation of neuron death. Limits the production of reactive oxygen species (ROS). Regulates cyclin-E during neuronal apoptosis. In collaboration with CHPF isoform 2, may enhance cell viability and protect cells from oxidative stress. -

Mirna and Parkinson's Disease Protein PARK2
open access www.bioinformation.net Editorial Volume 9(8) Molecule of the month: miRNA and Parkinson’s disease protein PARK2 Paul Shapshak1, 2 1Divsion of Infectious Disease and International Health, Department of Medicine and Department of Psychiatry and Behavioral Medicine, USF Morsani School of Medicine, Tampa General Hospital, 1 Tampa Gen Circle, Room G318, Tampa FL 33606; 2Deputy Chief Editor, Bioinformation; Paul Shapshak - Email: [email protected] Received April 07, 2013; Accepted April 07, 2013; Published April 30, 2013 Parkinson’s disease is generally associated with aging, i.e. Up-regulation, beige Regulation, purple Co-expression, brown elderly individuals. We are gaining many deep insights into Physical Interaction, turquoise dotted Predicted Protein neuropathogenesis in Parkinson’s disease and there are many Interaction, and mauve dotted Predicted TFactor Regulation publications on this topic; we briefly mention a few as they (GenePro SA Biosciences, http://www.sabiosciences.com/). relate to the increasingly studied miRNAs. Parkinson’s disease results from protein inclusions or Lewy bodies and destruction of (mid-brain) substantia nigra pars compacta neurons that are dopaminergic. A novel approach to investigate molecular processes in human diseases as well as in normal control and function is through the study of miRNAs. miRNAs are increasingly associated with many diseases and in Parkinson's disease [1, 2]. Figure 2: Network of input proteins from figure 1 (Park2, CASK, SNCAIP, HSPA4, STUB1, SIM2, CBL, LIMK1, PACRG, SEPT5, PSMA7, BAG5) and showing additional neighbors of these proteins (up to 100 total). In this figure, line-colors and various interactions with other genes are red Down-regulation, green Up-regulation, beige Regulation, purple Co-expression, Figure 1: Network of input protein (Park2) with immediate brown Physical Interaction, turquoise dotted Predicted Protein interactive proteins.