Jacquelyn S. Fetrow
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dickinson College Theta Adds 101 St Chapter a Small Liberal Arts College in Carlisle, PA Is the Home of Theta's One Hundred and First College Chapter
Ofd West, historicfocaf point on the campus al Dickinson. Dickinson College Theta adds 101 st chapter A small liberal arts college in Carlisle, PA is the home of Theta's one hundred and first college chapter. Founded in 1773 and related to the Methodist Church, Dickinson College is highly regarded academically with an admission standard rated "highly competitive." The campus at Dickinson is on 52 acres and buildings are in the Georgian architectural tradition. Epsilon Lambda joins four other Theta chapters in District VI which includes the State of Pennsylvania: Penn · State University, University of Pittsburgh, Carnegie-Mellon University, and Allegheny College. Charter members of Epsilon Lambda are: Nicole Anagnoste, Wyndmoor, PA; Wendy Beck, Pittsburgh; Winslow Bouscaren, Baltimore; Liza Chase, Golden's Bridge, NY; Sally Cochran, Jenkintown, PA; Alison Copley, Souderton, PA; Linda Coyne, Westfield, NJ; 1ennifer DeBerdine, Quarryville, PA; Alison Dickson, St. Michael's, MD; Robin Endicott, Belleplain, NJ; Beth Esler, Allentown, PA; Jane Fitzpatrick, Rumson, NJ; Robin Frabizio, Oakli.urst, NJ; Deborah Friend, Martinsville, NJ; Beth Gitlin, Butler, PA; Carrie Goodman, Baltimore; Marcy Grove, Midland, VA; Wendy Harkins, Exxon, PA; Alison Harkless, Altonna, PA; Anne Helmreich, Meadville, PA; Linda Janis, New Canaan, CT; Sarah Locke, Michigan City, IN; Carol Lookhoof, Morris Plains, · NJ; Valerie Ludlum, Ossining, NY; Michele McDonald, Inverness, IL; Diana Instaffation team on top .row f. tor. Carol Brehman, GVP Coffege; Miller, Hagerston, MD; Marlena Moors; Haddon Lissa Bradford, Grand President; Lynn Davis, G VP Service and lower row Judy Alexander, Grand Council Member-at-Large and Ann Heights, NJ; Nancy Oppenheimer, Wesi:lake Village, Gradwohl, Resident Counselor. -

2011 Annual Report of the Georgia Research Alliance Not Every Day Brings a Major Win, but in 2011, Many Did
2011 AnnuAl report of the GeorGiA reseArch AlliAnce not every day brings a major win, but in 2011, many did. An advance in science, a venture investment, the growth of a company – these kinds of developments, all connected in some way to the catalytic efforts of the Georgia research Alliance, signaled that Georgia’s technology-rich economy is alive and growing. And that’s good news. In GRA, Georgia has a game Beginning in July, GRA partnered These and other 2011 events Competition among states for plan for building the science- with the Georgia Department and activities represent a economic development and job and technology-driven of Economic Development to foundation for future creation is as intense as ever. economy of tomorrow. And determine how the state’s seven advances, investments and Every advantage matters. GRA in 2011, that game plan was Centers of Innovation, which economic growth. As such, bolsters Georgia’s competitive strengthened when newly help grow strategic industries, they are a reminder that it advantage by expanding elected Governor Nathan Deal can maximize their potential. pays to make every day count. university R&D capacity and moved to align more closely And the Georgia Cancer adding the fuel needed to several of Georgia’s greatest Coalition, the state’s signature launch new enterprises. assets for creating, expanding initiative for cancer research and attracting companies and care, was moved under the that foster high-wage jobs. GRA umbrella. the events of 2011 | JAnuArY – feBruArY Jan 1 Expert in autism named GRA Eminent Scholar Jan 11 Collaboration aims to develop new treatment for ovarian cancer Georgia Tech researchers have Meg Buscema proposed a filtration system that could potentially remove Billy Howard Jan 4 free-floating cancer cells that cause secondary tumors. -

Catalog 2008-09
DePauw University Catalog 2008-09 Preamble ...............................................................................................2 Section I: The University .....................................................................3 Section II: Graduation Requirements ...................................................8 Section III: Majors, Minors, Courses .................................................14 School of Music......................................................................18 College of Liberal Arts ...........................................................30 Graduate Programs in Education..........................................136 Section IV: Academic Policies .........................................................138 Section V: The DePauw Experience.................................................159 Section VI Campus Living ...............................................................176 Section VII: Admission, Expenses, Financial Aid ...........................184 Section VIII: University Personnel ..................................................196 This is a PDF copy of the official DePauw University Catalog, 2008-09, which is available at http://www.depauw.edu/catalog. This reproduction was created on September 15, 2008. Contact: Dr. Ken Kirkpatrick Registrar DePauw University 313 S. Locust St. Greencastle, IN 46135 [email protected] 765-658-4141 Preamble to the Catalog Accuracy of Catalog Information Every effort has been made to ensure that information in this catalog is accurate at the time of publication. -

For Ligand- Binding Site Prediction and Functional Annotation
A threading-based method (FINDSITE) for ligand- binding site prediction and functional annotation Michal Brylinski and Jeffrey Skolnick* Center for the Study of Systems Biology, School of Biology, Georgia Institute of Technology, 250 14th Street NW, Atlanta, GA 30318 Edited by Harold A. Scheraga, Cornell University, Ithaca, NY, and approved November 19, 2007 (received for review August 15, 2007) The detection of ligand-binding sites is often the starting point for Connolly surface (21) and then reranks the identified pockets by protein function identification and drug discovery. Because of the degree of conservation of identified surface residues. inaccuracies in predicted protein structures, extant binding pocket- A systematic analysis of known protein structures grouped detection methods are limited to experimentally solved structures. according to SCOP (22) reveals a general tendency of certain Here, FINDSITE, a method for ligand-binding site prediction and protein folds to bind substrates at a similar location, suggesting functional annotation based on binding-site similarity across that analogous or very distantly homologous proteins can have groups of weakly homologous template structures identified from common binding sites (11). If so, it should be possible to develop threading, is described. For crystal structures, considering a cutoff an approach for ligand-binding site identification that is less distance of4Åasthehitcriterion, the success rate is 70.9% for sensitive than pocket-detection methods to distortions in the identifying -

Daisuke Kihara, Ph.D. Professor of Biological Sciences and Computer Science Purdue University
Updated on June 21, 2020 Daisuke Kihara, Ph.D. Professor of Biological Sciences and Computer Science Purdue University Contact Information Purdue University Department of Biological Sciences and Computer Science West Lafayette, IN, 47907 Office: Hockmyer 229 Tel: (765) 496-2284 E-mail: [email protected] https://www.bio.purdue.edu/People/faculty_dm/directory.php?refID=166 https://www.cs.purdue.edu/people/faculty/dkihara/ http://kiharalab.org (Lab) Education 1999 Ph.D. (Science) in Bioinformatics Kyoto University, Faculty of Science, Japan, Advisor: Minoru Kanehisa 1996 M.S. in Bioinformatics Kyoto University, Faculty of Science, Japan 1994 B.S. in Interdisciplinary Science The University of Tokyo, College of Arts and Sciences, Japan Positions held 2019.3-present Full member, Purdue University Center for Cancer Research 2014.8-present Full Professor 2018.3-present Adjunct Professor, University of Cincinnati, Department of Pediatrics 2015.1-2015.8 Visiting Scientist, Eli Lilly, Indianapolis 2009.8-2014.8 Associate professor 2003.8-2009.8 tenure-track Assistant professor Purdue University, West Lafayette, Indiana Department of Biological Sciences/Computer Sciences (joint appointment) 2002.9-2003.7 Senior Postdoctoral Research Associate Advisor: Jeffrey Skolnick Buffalo Center of Excellence in Bioinformatics, Buffalo, NY, USA 1999-2002.9 Postdoctoral Research Associate Advisor: Jeffrey Skolnick Donald Danforth Plant Science Center, St. Louis, MO, USA 1998-1999 Research Assistant Advisor: Minoru Kanehisa Bioinformatics Center, Institute for -

Tertiary Structure Predictions on a Comprehensive Benchmark of Medium to Large Size Proteins
Biophysical Journal Volume 87 October 2004 2647–2655 2647 Tertiary Structure Predictions on a Comprehensive Benchmark of Medium to Large Size Proteins Yang Zhang and Jeffrey Skolnick Center of Excellence in Bioinformatics, University at Buffalo, Buffalo, New York 14203 ABSTRACT We evaluate tertiary structure predictions on medium to large size proteins by TASSER, a new algorithm that assembles protein structures through rearranging the rigid fragments from threading templates guided by a reduced Ca and side-chain based potential consistent with threading based tertiary restraints. Predictions were generated for 745 proteins 201– 300 residues in length that cover the Protein Data Bank (PDB) at the level of 35% sequence identity. With homologous proteins excluded, in 365 cases, the templates identified by our threading program, PROSPECTOR_3, have a root-mean-square deviation (RMSD) to native , 6.5 A˚ , with .70% alignment coverage. After TASSER assembly, in 408 cases the best of the top five full-length models has a RMSD , 6.5 A˚ . Among the 745 targets are 18 membrane proteins, with one-third having a predicted RMSD , 5.5 A˚ . For all representative proteins less than or equal to 300 residues that have corresponding multiple NMR structures in the Protein Data Bank, 20% of the models generated by TASSER are closer to the NMR structure centroid than the farthest individual NMR model. These results suggest that reasonable structure predictions for nonhomologous large size proteins can be automatically generated on a proteomic scale, and the application of this approach to structural as well as functional genomics represent promising applications of TASSER. INTRODUCTION The protein structure prediction problem, that is, deducing a RMSD , 7A˚ in the top five models (Simons et al., 2001). -

College/University Visit Clusters
COLLEGE/UNIVERSITY VISIT CLUSTERS The groupings of colleges and universities below are by no means exhaustive; these ideas are meant to serve as good starting points when beginning a college search. Happy travels! BOSTON/RHODE ISLAND AREA Large: Boston University University of Massachusetts at Boston Northeastern University Medium: Bentley University (business focus) Boston College Brandeis University Brown University Bryant College (business focus) Harvard University Massachusetts Institute of Technology Providence College University of Massachusetts at Lowell University of Rhode Island Suffolk University Small: Babson College (business focus) Emerson College Olin College Rhode Island School of Design (art school) Salve Regina University Simmons College (all women) Tufts University Wellesley College (all women) Wheaton College CENTRAL/WESTERN MASSACHUSETTS Large: University of Massachusetts at Amherst/Lowell Medium: College of the Holy Cross Worcester Polytechnic Institute Small: Amherst College Clark University Hampshire College Mount Holyoke College (all women) Smith College (all women) Westfield State University Williams College CONNECTICUT Large: University of Connecticut Medium: Fairfield University Quinnipiac University Yale University Small: Connecticut College Trinity College Wesleyan University NORTHERN NEW ENGLAND Large: University of New Hampshire University of Vermont Medium: Dartmouth College Middlebury College Small: Bates College Bennington College Bowdoin College Colby College College of the Atlantic Saint Anselm College -

Colleges & Universities
Bishop Watterson High School Students Have Been Accepted at These Colleges and Universities Art Institute of Chicago Fordham University Adrian College University of Cincinnati Franciscan University of Steubenville University of Akron Cincinnati Art Institute Franklin and Marshall College University of Alabama The Citadel Franklin University Albion College Claremont McKenna College Furman University Albertus Magnus College Clemson University Gannon University Allegheny College Cleveland Inst. Of Art George Mason University Alma College Cleveland State University George Washington University American Academy of Dramatic Arts Coastal Carolina University Georgetown University American University College of Charleston Georgia Southern University Amherst College University of Colorado at Boulder Georgia Institute of Technology Anderson University (IN) Colorado College University of Georgia Antioch College Colorado State University Gettysburg College Arizona State University Colorado School of Mines Goshen College University of Arizona Columbia College (Chicago) Grinnell College (IA) University of Arkansas Columbia University Hampshire College (MA) Art Academy of Cincinnati Columbus College of Art & Design Hamilton College The Art Institute of California-Hollywood Columbus State Community College Hampton University Ashland University Converse College (SC) Hanover College (IN) Assumption College Cornell University Hamilton College Augustana College Creighton University Harvard University Aurora University University of the Cumberlands Haverford -
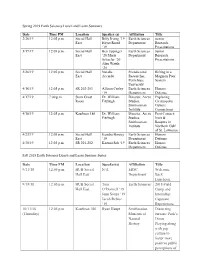
Past ERSC Seminar Series 1992-Current
Spring 2019 Earth Sciences Lunch and Learn Seminars Date Time PM Location Speaker (s) Affiliation Title 2/26/19 12:05 p.m. Social Hall Billy Irving ’19 Earth Sciences Senior East Hayat Rasul Department Research ‘19 Presentations 3/19/19 12:05 p.m. Social Hall Ben Eppinger Earth Sciences Junior East ’20 Maria Department Research Schaefer ’20 Presentations Alex Wattle ‘20 3/26/19 12:05 p.m. Social Hall Natalie Postdoctoral Rifting in a East Accardo Researcher, Magman Poor Penn State System University 4/16/19 12:05 p.m. SR 202-203 Allison Curley Earth Sciences Honors ‘19 Department Defense 4/17/19 7:00 p.m. Stern Great Dr. William Director, Arctic Exploring Room Fitzhugh Studies, Circumpolar Smithsonian Culture Institute Connections 4/18/19 12:05 p.m. Kaufman 186 Dr. William Director, Arctic First Contact: Fitzhugh Studies, Inuit & Smithsonian Basques in Institute Northern Gulf of St. Lawrence 4/23/19 12:05 p.m. Social Hall Kendra Bonsey Earth Sciences Honors East ‘19 Department Defense 4/30/19 12:05 p.m. SR 201-202 Karuna Sah ‘19 Earth Sciences Honors Department Defense Fall 2018 Earth Sciences Lunch and Learn Seminar Series Date Time PM Location Speaker(s) Affiliation Title 9/11/18 12:05 p.m. HUB Social N/A ERSC Welcome Hall East Department Back Luncheon 9/18/18 12:05 p.m. HUB Social Tom Earth Sciences 2018 Field Hall East O’Donnell ‘19 Camp and Jenn Souza ‘19 Internship Jacob Rebisz Capstone ‘19 Experiences 10/11/18 12:05 p.m. -

Jacquelyn S. Fetrow Executive Resume Office of the President Work Email: [email protected] Library and Administration Building Office Phone: 610-921-7600 N
Jacquelyn S. Fetrow Executive Resume Office of the President Work Email: [email protected] Library and Administration Building Office Phone: 610-921-7600 N. 13th and Bern Streets, P.O. Box 15234 Reading, PA 19612 Current Position: President and Professor of Chemistry June 2017-present Education Ph.D. Biological Chemistry, Penn State, December, 1986 B.S. Biochemistry, Albright College, May, 1982 Professional Experience University of Richmond, Richmond, VA Provost and Vice President for Academic Affairs July 2014-Dec 2016 Professor of Chemistry July 2014-May 2017 Wake Forest University, Winston-Salem, NC Dean, Wake Forest College January 2009-June 2014 Reynolds Professor of Computational Biophysics August 2003-June 2014 Concurrent appointments: Affiliated Faculty, Wake Forest School of Medicine Cancer Center. Department of Biochemistry, Program in Molecular Medicine, Program in Molecular Genetics and Genomics, School of Biomedical Engineering and Science (SBES, joint program between Virginia Tech and Wake Forest) Director, Graduate Track in Structural and Computational Biophysics 2005-2008 GeneFormatics, Inc., San Diego, CA Co-founder, Chief Scientific Officer and Director May 1999-January, 2003 The Scripps Research Institute, La Jolla, CA Associate Professor, Department of Molecular Biology April 1998-May 1999 Visiting Scientist, Department of Molecular Biology (laboratory of Jeffrey Skolnick) January-December 1997 The University at Albany, SUNY, Albany, NY Associate Professor (with tenure), Department of Biological Sciences September 1995-April 1998 Assistant Professor, Department of Biological Sciences January 1990-August 1995 Concurrent/joint appointments: Department of Chemistry, Department of Biomedical Sciences, School of Public Health, Department of Biomedical Science, School of Public Health Postdoctoral Fellowships The Whitehead Institute for Biomedical Research, MIT, Cambridge, MA (Advisor: Peter S. -
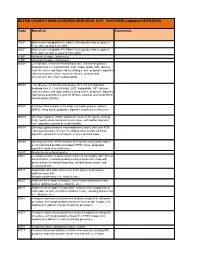
SETON CHARITY NON-COVERED SERVICES EFF: 01/01/2020 (Updated 05/05/2020)
SETON CHARITY NON-COVERED SERVICES EFF: 01/01/2020 (updated 05/05/2020) Code Narrative Comments 3051F Most recent hemoglobin A1c (HbA1c) level greater than or equal to 7.0% and less than 8.0% (DM) 3052F Most recent hemoglobin A1c (HbA1c) level greater than or equal to 8.0% and less than or equal to 9.0% (DM) 4185F Histamine therapy - intravenous 4186F Histamine therapy - intravenous 0002M Liver disease, ten biochemical assays (ALT, A2-macro- globulin, apolipoprotein A-1, total bilirubin, GGT, hapto- globin, AST, glucose, total cholesterol and triglycerides) utilizing serum, prognostic algorithm reported as quanti- tative scores for fibrosis, steatosis and alcoholic/non alco- holic steatohepatitis 0003M Liver disease, ten biochemical assays (ALT, A2-macroglobulin, apolipoprotein A-1, total bilirubin, GGT, haptoglobin, AST, glucose, total cholesterol and triglycerides) utilizing serum, prognostic algorithm reported as quantitative scores for fibrosis, steatosis and nonalcoholic steatohepatitis (NASH) 0004M Scoliosis, DNA analysis of 53 single nucleotide polymor- phisms (SNPs), using saliva, prognostic algorithm reported as a risk score 0006M Oncology (hepatic), mRNA expression levels of 161 genes, utilizing fresh hepatocellular carcinoma tumor tissue, with alpha-fetoprotein level, algorithm reported as a risk classifier 0007M Oncology (gastrointestinal neuroendocrine tumors), real- time PCR expression analysis of 51 genes, utilizing whole peripheral blood, algorithm reported as a nomogram of tumor disease index 0008M Oncology (breast), mRNA -

1 Depauw University Faculty Meeting Agenda December 7, 2015 1. Call
DePauw University Faculty Meeting Agenda December 7, 2015 1. Call to Order – 4 p.m. Union Building Ballroom The Chair welcomed everyone and make a few quick reminders: • Let’s continue to be inclusive in our conversations by always introducing ourselves when we speak. • If you’d like to speak please come to one of the microphones so everyone can hear you, depending on where folks are sitting the acoustics are great or NOT. • If you don’t like to be startled when your cell phone rings aloud, please check that it is silenced. 2. Verification of Quorum (86 for the fall) Jim Mills signaled that a quorum was reached at 4:05 p.m. 3. Faculty Remembrances for Catherine E. Fruhan Catherine E. Fruhan, Professor of Art History passed away unexpectedly November 20, 2015. She was a full-time faculty member at DePauw from 1984 through her passing in 2015. Anne Harris, Vice President for Academic Affairs and Professor of Art History wrote and read the remembrance found in Appendix A. 4. Faculty Remembrances for Glenn E. Welliver Glenn E. Welliver, Professor Emeritus of German was a full-time faculty member at DePauw from 1964 to 1999. Glenn passed away on November 8, 2015. Professor Emeritus of Romance Languages, James Rambo wrote the remembrance found in Appendix B. Alejandro Puga, Associate Professor of Spanish and Chair of the Department of Modern Languages read the remembrance. 5. Consent Agenda There were no requests to move anything from the consent agenda to a regular item of business. The consent agenda was approved.