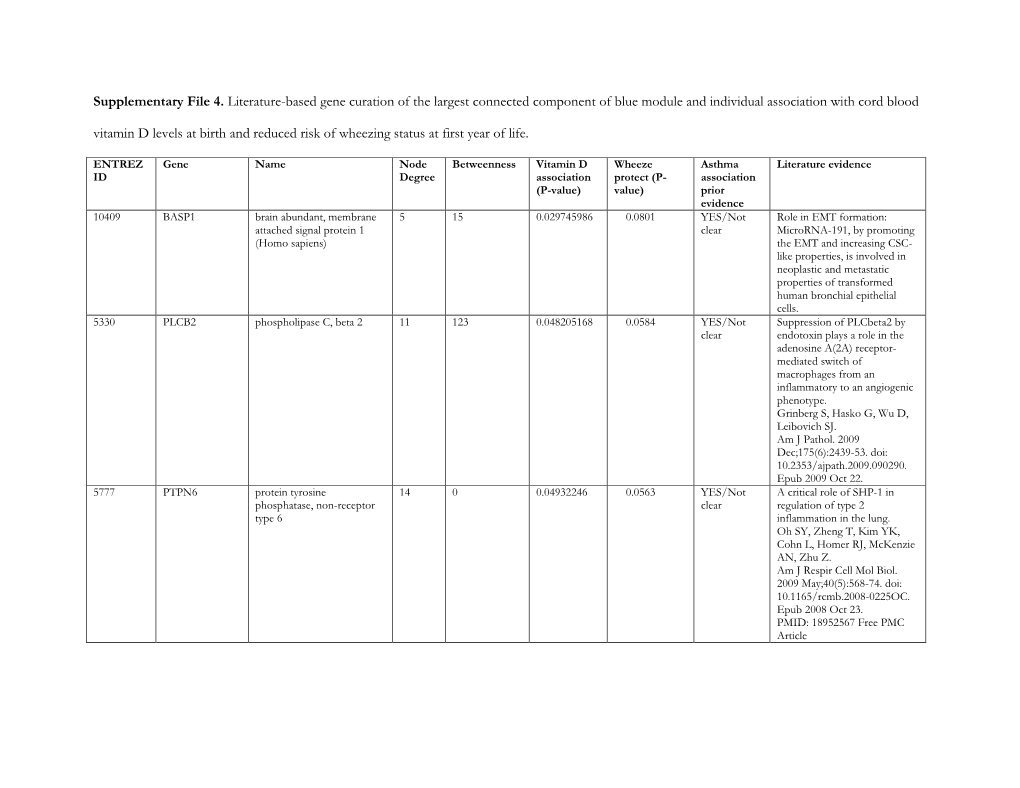
Supplementary File 4. Literature-Based Gene Curation of the Largest Connected Component of Blue Module and Individual Associati
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

IQSEC2 Antibody Cat
IQSEC2 Antibody Cat. No.: 8011 Western blot analysis of IQSEC2 in SK-N-SH cell lysate with IQSEC2 antibody at 1 μg/ml. Immunohistochemistry of IQSEC2 in mouse brain tissue with IQSEC2 antibody at 5 μg/ml. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse, Rat IQSEC2 antibody was raised against an 18 amino acid peptide near the carboxy terminus of human IQSEC2. IMMUNOGEN: The immunogen is located within amino acids 1380 - 1430 of IQSEC2. TESTED APPLICATIONS: ELISA, IHC-P, WB IQSEC2 antibody can be used for detection of IQSEC2 by Western blot at 1 - 2 μg/ml. Antibody can also be used for immunohistochemistry starting at 5 μg/mL. APPLICATIONS: Antibody validated: Western Blot in human samples and Immunohistochemistry in mouse samples. All other applications and species not yet tested. September 27, 2021 1 https://www.prosci-inc.com/iqsec2-antibody-8011.html IQSEC2 antibody is human, mouse and rat reactive. At least two isoforms of IQSEC2 are SPECIFICITY: known to exist; this antibody will only detect the larger isoform. IQSEC2 antibody is predicted to not cross-react with IQSEC1. POSITIVE CONTROL: 1) Cat. No. 1220 - SK-N-SH Cell Lysate Predicted: 104, 133, 141, 164 kDa PREDICTED MOLECULAR WEIGHT: Observed: 140 kDa Properties PURIFICATION: IQSEC2 antibody is affinity chromatography purified via peptide column. CLONALITY: Polyclonal ISOTYPE: IgG CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: IQSEC2 antibody is supplied in PBS containing 0.02% sodium azide. CONCENTRATION: 1 mg/mL IQSEC2 antibody can be stored at 4˚C for three months and -20˚C, stable for up to one STORAGE CONDITIONS: year. -

Tristetraprolin Limits Inflammatory Cytokine Production in Tumor-Associated Macrophages in an Mrna Decay− Independent Manner
Published OnlineFirst July 16, 2015; DOI: 10.1158/0008-5472.CAN-15-0205 Cancer Microenvironment and Immunology Research Tristetraprolin Limits Inflammatory Cytokine Production in Tumor-Associated Macrophages in an mRNA Decay–Independent Manner Franz Kratochvill1,2, Nina Gratz1, Joseph E. Qualls1,2, Lee-Ann Van De Velde1,2, Hongbo Chi2, Pavel Kovarik3, and Peter J. Murray1,2 Abstract Tristetraprolin (TTP) is an inducible zinc finger AU-rich (TAM). However, TTP's effects on AU-rich mRNA stability RNA-binding protein essential for enforcing degradation of were negligible and limited by constitutive p38a MAPK activ- mRNAs encoding inflammatory chemokines and cytokines. ity, which was the main driver of proinflammatory cytokine Most studies on TTP center on the connection between mRNA production in TAMs at the posttranscriptional level. Instead, half-life and inflammatory output, because loss of TTP ampli- elimination of TTP caused excessive protein production of fies inflammation by increasing the stability of AU-rich inflammatory mediators, suggesting TTP-dependent transla- mRNAs. Here, we focused on how TTP controls cytokine and tional suppression of AU-rich mRNAs. Manipulation of the chemokine production in the nonresolving inflammation of p38a–TTP axis in macrophages has significant effects on the cancer using tissue-specific approaches. In contrast with mod- growth of tumors and therefore represents a means to mani- el in vitro macrophage systems, we found constitutive TTP pulate inflammation in the tumor microenvironment. Cancer expression in late-stage tumor-associated macrophages Res; 75(15); 1–11. Ó2015 AACR. Introduction TLR signaling phosphorylates TTP thereby blocking its function and sustaining TNF output (9, 10). -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Integrative Differential Expression and Gene Set Enrichment Analysis Using Summary Statistics for Scrna-Seq Studies
ARTICLE https://doi.org/10.1038/s41467-020-15298-6 OPEN Integrative differential expression and gene set enrichment analysis using summary statistics for scRNA-seq studies ✉ Ying Ma 1,7, Shiquan Sun 1,7, Xuequn Shang2, Evan T. Keller 3, Mengjie Chen 4,5 & Xiang Zhou 1,6 Differential expression (DE) analysis and gene set enrichment (GSE) analysis are commonly applied in single cell RNA sequencing (scRNA-seq) studies. Here, we develop an integrative 1234567890():,; and scalable computational method, iDEA, to perform joint DE and GSE analysis through a hierarchical Bayesian framework. By integrating DE and GSE analyses, iDEA can improve the power and consistency of DE analysis and the accuracy of GSE analysis. Importantly, iDEA uses only DE summary statistics as input, enabling effective data modeling through com- plementing and pairing with various existing DE methods. We illustrate the benefits of iDEA with extensive simulations. We also apply iDEA to analyze three scRNA-seq data sets, where iDEA achieves up to five-fold power gain over existing GSE methods and up to 64% power gain over existing DE methods. The power gain brought by iDEA allows us to identify many pathways that would not be identified by existing approaches in these data. 1 Department of Biostatistics, University of Michigan, Ann Arbor, MI 48109, USA. 2 School of Computer Science, Northwestern Polytechnical University, Xi’an, Shaanxi 710072, P.R. China. 3 Department of Urology, University of Michigan, Ann Arbor, MI 48109, USA. 4 Department of Human Genetics, University of Chicago, Chicago, IL 60637, USA. 5 Section of Genetic Medicine, Department of Medicine, University of Chicago, Chicago, IL 60637, USA. -

Integrating Text Mining, Data Mining, and Network Analysis for Identifying
Jurca et al. BMC Res Notes (2016) 9:236 DOI 10.1186/s13104-016-2023-5 BMC Research Notes RESEARCH ARTICLE Open Access Integrating text mining, data mining, and network analysis for identifying genetic breast cancer trends Gabriela Jurca1, Omar Addam1, Alper Aksac1, Shang Gao2, Tansel Özyer3, Douglas Demetrick4 and Reda Alhajj1,5* Abstract Background: Breast cancer is a serious disease which affects many women and may lead to death. It has received considerable attention from the research community. Thus, biomedical researchers aim to find genetic biomarkers indicative of the disease. Novel biomarkers can be elucidated from the existing literature. However, the vast amount of scientific publications on breast cancer make this a daunting task. This paper presents a framework which investigates existing literature data for informative discoveries. It integrates text mining and social network analysis in order to identify new potential biomarkers for breast cancer. Results: We utilized PubMed for the testing. We investigated gene–gene interactions, as well as novel interactions such as gene-year, gene-country, and abstract-country to find out how the discoveries varied over time and how overlapping/diverse are the discoveries and the interest of various research groups in different countries. Conclusions: Interesting trends have been identified and discussed, e.g., different genes are highlighted in relation- ship to different countries though the various genes were found to share functionality. Some text analysis based results have been validated against results from other tools that predict gene–gene relations and gene functions. Keywords: Breast cancer, Data mining, Text mining, Network analysis Background reducing the number of molecules to consider as poten- Introduction tial biomarkers. -

NF-Y Inactivation Causes Atypical Neurodegeneration Characterized by Ubiquitin and P62 Accumulation and Endoplasmic Reticulum Disorganization
ARTICLE Received 25 Jul 2013 | Accepted 30 Jan 2014 | Published 25 Feb 2014 DOI: 10.1038/ncomms4354 NF-Y inactivation causes atypical neurodegeneration characterized by ubiquitin and p62 accumulation and endoplasmic reticulum disorganization Tomoyuki Yamanaka1,2,3,4, Asako Tosaki2,4, Masaru Kurosawa1,2,3,4, Gen Matsumoto1,2,3,4, Masato Koike5, Yasuo Uchiyama5, Sankar N. Maity6, Tomomi Shimogori3, Nobutaka Hattori7 & Nobuyuki Nukina1,2,3,4 Nuclear transcription factor-Y (NF-Y), a key regulator of cell-cycle progression, often loses its activity during differentiation into nonproliferative cells. In contrast, NF-Y is still active in mature, differentiated neurons, although its neuronal significance remains obscure. Here we show that conditional deletion of the subunit NF-YA in postmitotic mouse neurons induces progressive neurodegeneration with distinctive ubiquitin/p62 pathology; these proteins are not incorporated into filamentous inclusion but co-accumulated with insoluble membrane proteins broadly on endoplasmic reticulum (ER). The degeneration also accompanies drastic ER disorganization, that is, an aberrant increase in ribosome-free ER in the perinuclear region, without inducing ER stress response. We further perform chromatin immunoprecipitation and identify several NF-Y physiological targets including Grp94 potentially involved in ER disorganization. We propose that NF-Y is involved in a unique regulation mechanism of ER organization in mature neurons and its disruption causes previously undescribed novel neuropathology accompanying abnormal ubiquitin/p62 accumulation. 1 Department of Neuroscience for Neurodegenerative Disorders, Juntendo University Graduate School of Medicine, 2-1-1 Hongo, Bunkyo-ku, Tokyo 113-8421, Japan. 2 Laboratory for Structural Neuropathology, RIKEN Brain Science Institute, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan. -

Westminsterresearch ZFP36 Proteins and Mrna Targets in B Cell
WestminsterResearch http://www.westminster.ac.uk/westminsterresearch ZFP36 proteins and mRNA targets in B cell malignancies Alcaraz, A. This is an electronic version of a PhD thesis awarded by the University of Westminster. © Miss Amor Alcaraz, 2015. The WestminsterResearch online digital archive at the University of Westminster aims to make the research output of the University available to a wider audience. Copyright and Moral Rights remain with the authors and/or copyright owners. Whilst further distribution of specific materials from within this archive is forbidden, you may freely distribute the URL of WestminsterResearch: ((http://westminsterresearch.wmin.ac.uk/). In case of abuse or copyright appearing without permission e-mail [email protected] ZFP36 proteins and mRNA targets in B cell malignancies Maria del Amor Alcaraz-Serrano A Thesis submitted in partial fulfilment of the requirements of the University of Westminster for the degree of Doctor of Philosophy September 2015 Abstract The ZFP36 proteins are a family of post-transcriptional regulator proteins that bind to adenine uridine rich elements (AREs) in 3’ untranslated (3’UTR) regions of mRNAs. The members of the human family, ZFP36L1, ZFP36L2 and ZFP36 are able to degrade mRNAs of important cell regulators that include cytokines, cell signalling proteins and transcriptional factors. This project investigated two proposed targets for the protein family that have important roles in B cell biology, BCL2 and CD38 mRNAs. BCL2 is an anti-apoptotic protein with key roles in cell survival and carcinogenesis; CD38 is a membrane protein differentially expressed in B cells and with a prognostic value in B chronic lymphocytic leukaemia (B-CLL), patients positive for CD38 are considered to have a poor prognosis. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Expressed Gene Fusions As Frequent Drivers of Poor Outcomes in Hormone Receptor–Positive Breast Cancer
Published OnlineFirst December 14, 2017; DOI: 10.1158/2159-8290.CD-17-0535 RESEARCH ARTICLE Expressed Gene Fusions as Frequent Drivers of Poor Outcomes in Hormone Receptor–Positive Breast Cancer Karina J. Matissek1,2, Maristela L. Onozato3, Sheng Sun1,2, Zongli Zheng2,3,4, Andrew Schultz1, Jesse Lee3, Kristofer Patel1, Piiha-Lotta Jerevall2,3, Srinivas Vinod Saladi1,2, Allison Macleay3, Mehrad Tavallai1,2, Tanja Badovinac-Crnjevic5, Carlos Barrios6, Nuran Beşe7, Arlene Chan8, Yanin Chavarri-Guerra9, Marcio Debiasi6, Elif Demirdögen10, Ünal Egeli10, Sahsuvar Gökgöz10, Henry Gomez11, Pedro Liedke6, Ismet Tasdelen10, Sahsine Tolunay10, Gustavo Werutsky6, Jessica St. Louis1, Nora Horick12, Dianne M. Finkelstein2,12, Long Phi Le2,3, Aditya Bardia1,2, Paul E. Goss1,2, Dennis C. Sgroi2,3, A. John Iafrate2,3, and Leif W. Ellisen1,2 ABSTRACT We sought to uncover genetic drivers of hormone receptor–positive (HR+) breast cancer, using a targeted next-generation sequencing approach for detecting expressed gene rearrangements without prior knowledge of the fusion partners. We identified inter- genic fusions involving driver genes, including PIK3CA, AKT3, RAF1, and ESR1, in 14% (24/173) of unselected patients with advanced HR+ breast cancer. FISH confirmed the corresponding chromo- somal rearrangements in both primary and metastatic tumors. Expression of novel kinase fusions in nontransformed cells deregulates phosphoprotein signaling, cell proliferation, and survival in three- dimensional culture, whereas expression in HR+ breast cancer models modulates estrogen-dependent growth and confers hormonal therapy resistance in vitro and in vivo. Strikingly, shorter overall survival was observed in patients with rearrangement-positive versus rearrangement-negative tumors. Cor- respondingly, fusions were uncommon (<5%) among 300 patients presenting with primary HR+ breast cancer. -

Transcriptomic Profiling of Ca Transport Systems During
cells Article Transcriptomic Profiling of Ca2+ Transport Systems during the Formation of the Cerebral Cortex in Mice Alexandre Bouron Genetics and Chemogenomics Lab, Université Grenoble Alpes, CNRS, CEA, INSERM, Bâtiment C3, 17 rue des Martyrs, 38054 Grenoble, France; [email protected] Received: 29 June 2020; Accepted: 24 July 2020; Published: 29 July 2020 Abstract: Cytosolic calcium (Ca2+) transients control key neural processes, including neurogenesis, migration, the polarization and growth of neurons, and the establishment and maintenance of synaptic connections. They are thus involved in the development and formation of the neural system. In this study, a publicly available whole transcriptome sequencing (RNA-Seq) dataset was used to examine the expression of genes coding for putative plasma membrane and organellar Ca2+-transporting proteins (channels, pumps, exchangers, and transporters) during the formation of the cerebral cortex in mice. Four ages were considered: embryonic days 11 (E11), 13 (E13), and 17 (E17), and post-natal day 1 (PN1). This transcriptomic profiling was also combined with live-cell Ca2+ imaging recordings to assess the presence of functional Ca2+ transport systems in E13 neurons. The most important Ca2+ routes of the cortical wall at the onset of corticogenesis (E11–E13) were TACAN, GluK5, nAChR β2, Cav3.1, Orai3, transient receptor potential cation channel subfamily M member 7 (TRPM7) non-mitochondrial Na+/Ca2+ exchanger 2 (NCX2), and the connexins CX43/CX45/CX37. Hence, transient receptor potential cation channel mucolipin subfamily member 1 (TRPML1), transmembrane protein 165 (TMEM165), and Ca2+ “leak” channels are prominent intracellular Ca2+ pathways. The Ca2+ pumps sarco/endoplasmic reticulum Ca2+ ATPase 2 (SERCA2) and plasma membrane Ca2+ ATPase 1 (PMCA1) control the resting basal Ca2+ levels. -

Integrating Protein Copy Numbers with Interaction Networks to Quantify Stoichiometry in Mammalian Endocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Integrating protein copy numbers with interaction networks to quantify stoichiometry in mammalian endocytosis Daisy Duan1, Meretta Hanson1, David O. Holland2, Margaret E Johnson1* 1TC Jenkins Department of Biophysics, Johns Hopkins University, 3400 N Charles St, Baltimore, MD 21218. 2NIH, Bethesda, MD, 20892. *Corresponding Author: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Proteins that drive processes like clathrin-mediated endocytosis (CME) are expressed at various copy numbers within a cell, from hundreds (e.g. auxilin) to millions (e.g. clathrin). Between cell types with identical genomes, copy numbers further vary significantly both in absolute and relative abundance. These variations contain essential information about each protein’s function, but how significant are these variations and how can they be quantified to infer useful functional behavior? Here, we address this by quantifying the stoichiometry of proteins involved in the CME network. We find robust trends across three cell types in proteins that are sub- vs super-stoichiometric in terms of protein function, network topology (e.g.