Best Practices for Research Reproducibility.Pdf
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Employer Reference
Updated: May, 2012 WebDT Signage Player 4.X Supported Media & Performance Guideline Table of Contents Preface ....................................................................................................................................................................................................................................... 1 Media Supported Guideline ...................................................................................................................................................................................................... 2 Video Performance Guideline ................................................................................................................................................................................................... 8 Glossary ................................................................................................................................................................................................................................... 12 Preface This document outlines supported media formats and video performance reference on the WebDT Signage Appliance Version 4.X. We strongly recommend you to read related information for the SA model you have purchased before you start to play your media contents. 1 Updated: May, 2012 Media Supported Guideline i. Supported Formats Supported File Format Video Encoding Audio Encoding Remark File Extension MP4 .mp4 MPEG4 AAC, MP3 H.264 AAC, MP3 MOV .mov MP4 AAC, AMR Narrowband, IMA 4:1, PCM, Note: Apple lossless, MACE3-1, -

Audio File Types for Preservation and Access
AUDIO FILE TYPES FOR PRESERVATION AND ACCESS INTRODUCTION This resource guide identifies and compares audio file formats commonly used for preservation masters and access copies for both born digital and digitized materials. There are a wide range of audio file formats available, each with their own uses, considerations, and best practices. For more information about technical details see the glossary and additional resources linked at the end of this resource guide. For more information about audio files, view related items connected to this resource on the Sustainable Heritage Network in the “Audio Recordings” category. AUDIO FILE FORMATS FOR PRESERVATION MASTERS Lossless files (either uncompressed, or using lossless compression) are best used for preservation masters as they have the highest audio quality and fidelity, however, they produce the largest file sizes. Preservation Masters are generally not edited, or are minimally edited because their purpose is to serve as the most faithful copy of the original recording possible. WAV (Waveform Audio File Format) ● Uncompressed ● Proprietary (IBM), but very widely used ● Accessible on all operating systems and most audio software ● A variant called BWF (Broadcast Wave File Format) allows embedding of additional metadata sustainableheritagenetwork.org | [email protected] Center for Digital Scholarship and Curation | cdsc.libraries.wsu.edu Resource updated 3/14/2018 FLAC (Free Lossless Audio Codec) ● Compressed (lossless) ● Open format ● Accessible on all operating -

Research Data Management Good Practice Note
SMB-ARC 13/9/17 Research Data Management Good Practice Note Version: September 2017 RESEARCH DATA MANAGEMENT Good Practice Note Prepared by: CGIAR Internal Audit Unit Page 1 of 45 Version: September 2017 Table of contents FOREWORD ................................................................................................................................................... 3 1. INTRODUCTION ......................................................................................................................................... 4 1.1 What is Research Data? ...................................................................................................................... 4 1.2 Benefits of research data management.............................................................................................. 4 1.3 Potential risks associated with poor research data management ...................................................... 5 1.3.1 Risks related to Research Data Management, their sources and implications ........................... 6 2. State of RDM at CGIAR .............................................................................................................................. 6 2.1 CGIAR System requirements ............................................................................................................... 6 2.1.1 CGIAR Open Access and Data Management Policy ..................................................................... 6 2.1.2 CGIAR Principles on the Management of Intellectual Assets ..................................................... -

List of File Formats
Nicole Martin November 4, 2007 Digital Preservation - MIAP List of File Formats File Name: Advanced Audio Coding File Extension: .aac Creator: Collaboration between corporations approved by MPEG Creation Date: 1997 Media Type: Sound Format: Lossy Compression Notes: Developed to work as a part of MPEG-4, the AAC file format employs a very efficient form of compression meant to improve on the standard MP3. File Name: Advanced Authoring Format File Extension: .aaf Creator: Advanced Media Workflow Association Creation Date: 2000 Media Type: Moving Image Format: Uncompressed Notes: Created specifically for use in the post-production/editing environment to address interoperability issues. The AAF file format acts as a wrapper, is capable of storing metadata, and was designed to be versatile enough to withstand the myriad changes inherent in a production setting. File Name: Audio Interchange File Format File Extension: .aiff Creator: Electronic Arts Interchange and Apple Computer, Inc. Creation Date: 1988 Media Type: Sound Format: Uncompressed Notes: Standard Macintosh file format that is compatible with Windows (.aif) and often used by digital audio devices. Regular AIFF files are uncompressed, but compressed versions of the format (AIFF-C or AIFC) were developed to function with various codecs. File Name: Audio Video Interleave File Extension: .avi Creator: Microsoft Creation Date: 1992 Media Type: Moving Image Format: Container Notes: File most often created when DV files are imported from a camcorder to computer. File Name: Bitmap File Extension: .bmp Creator: IBM and Microsoft Creation Date: 1988 Media Type: Still Image Format: Compressed or Uncompressed Notes: Originally created for release with the Windows OS/2 operating system, BMPs are raster image files that are able to range from large, high quality files to small files of lesser quality. -

List of EPFL Recommended File Formats
List of EPFL Recommended File Formats Type of Data Sub Type Recommended File Formats for Sharing, Reuse and Long-Term Preservation Acceptable Formats (up to 10 year) Not Suitable for Preservation Dataset Tabular data with extensive metadata comma-separated values (CSV) file unicode UTF-8 (.csv) with CSV on the Web descriptive metadata plain text data, ASCII (.txt) Hierarchical Data Format version 5 HDF5 (.hdf5) Hypertext Mark-up Language (HTML) (.html) LaTeX (.tex) SPSS portable format (.por) Tabular data with minimal metadata comma-separated values (CSV) unicode UTF-8 file (.csv) eXtensible Mark-up Language (XML) text according to an appropriate MS Excel (xls, .xlsb) tab-delimited file (.tab) Document Type Definition (DTD) or schema (.xml) OpenDocument Spreadsheet (.ods) MS Excel (.xlsx) Structured Query Language (SQL) dump, preferably from an open tool (PostgreSQL, MariaDB) Text Textual data PDF/A (.pdf) widely-used proprietary formats, e.g. MS Word (.docx) PowerPoint (.pptx) MS Word (.doc) plain text, unicode UTF-8 (.txt) PDF with embedded forms PowerPoint (.ppt) Open Document Text (.odt, .odm) Rich Text Format (.rtf) LaTeX (.tex) Markdown (.md) HTML (.htm, .html), XHMTL 1.0 XML marked-up text (.xml), with specified schema Code Plain text formats (e.g. Matlab/Octave .m , R-project .R, Python .py , and so on) Text files for S-plus (.sdd) Matlab (.mat) Jupyter Notebook (.iypnb) Matlab .mat, should be saved in HDF format. R-project (.rdata) Rstudio (.rstudio), Rmarkdown (.rmd) NetCDF Multimedia Digital image data Raster : Raster : Vector -

Checklist for a Software Management Plan
Checklist for a Software Management Plan Michael Jackson (ed.) Version 0.2 10.5281/zenodo.1460504 12 October 2018 Please cite as: Michael Jackson (ed.) (12 October 2018). Checklist for a Software Management Plan (Version 0.2). Zenodo. doi:10.5281/zenodo.1460504. Web site: https://www.software.ac.uk/software- management-plans Introduction It is easy to concentrate on short-term issues when developing research software. Getting publications, collaboration with others and the demands of a daily research routine can all conspire to prevent proper planning for the development of research software. A Software Management Plan can help us to define a set of structures and goals to help us to understand what we are going to write, who it is for, how we will get it to them, how will it help them, and how we will assess whether it has helped them. They also help us to understand what processes, resources and infrastructure we need and how we can use these to meet our own goals, in the short, medium and long term. They also encourage us to think about the future of our software once our project or funding period ends, and what our plans for its long-term sustainability are. This checklist is intended to help researchers write software management plans. Use of this checklist The Software Sustainability Institute provides this checklist on an "as-is" basis, makes no warranties regarding any information provided within and disclaims liability for damages resulting from using this information. You are solely responsible for determining the appropriateness of any advice and guidance provided and assume any risks associated with your use of this advice and guidance. -

File Formats
Electronic Records Management Guidelines File Formats File Formats Summary Rapid changes in technology mean that file formats can become obsolete quickly and cause problems for your records management strategy. A long-term view and careful planning can overcome this risk and ensure that you can meet your legal and operational requirements. Legally, your records must be trustworthy, complete, accessible, legally admissible in court, and durable for as long as your approved records retention schedules require. For example, you can convert a record to another, more durable format (e.g., from a nearly obsolete software program to a text file) and that copy, as long as it is created in a trustworthy manner, is legally acceptable. The software in which a file is created usually uses a default format when the file is saved. This is indicated by the file name suffix (e.g., .PDF for portable document format). However, most software allows authors to select from a variety of formats when they save a file. For example, Microsoft Word allows the author to select document [DOC], Rich Text Format [RTF], or text [TXT], as well as other format options. Some software, such as Adobe Acrobat, is designed to convert files from one format to another. The format you choose will affect your long-term records management abilities. Legal Framework For more information on the legal framework you must consider when developing a file format policy refer to the Legal Framework chapter of these guidelines and the Minnesota State Archives’ Preserving and Disposing of Government Records1. Key Concepts As you consider the file format options available to you, you will need to be familiar with the following concepts: Proprietary, Non-Proprietary, Open-Source, and Open Standard File Formats File Types and their Associated Formats Preservation Options: Conversion and Migration Compression Importance of Planning 1 Minnesota Historical Society. -

A-PDF Djvu To
PDF DjVu to PDF utility User Documentation Note: This product is distributed on a ‘try -before -you -buy’ basis. All features described in this documentation are enabled. The registered version does not insert a watermark in your generated pdf documents. About A-PDF DjVu to PDF A-PDF DjVu to PDF is a fast, affordable way to batch convert DjVu documents (.DjVu) into professional-quality documents in the popular PDF file format. Its easy-to-use interface allows you to batch create PDF files which can be viewed on any computer with a PDF viewer. A-PDF DjVu to PDF supports font embedding, adding watermark, security, page number and multi-language. Released: September 2009 Page 1 of 14 Copyright © 2009 A-PDF.com - all rights reserved PDF DjVu to PDF utility User Documentation A-PDF DjVu to PDF lets you instantly convert DjVu Documents (.DjVu) into fully- formatted and professional-quality PDF file format. A-PDF DjVu to PDF retains the layout of the original DjVu document, and it supports all PDF file settings, such as PDF Compatibility, Auto-Rotate, Resolution, Compress settings of PDF document, Colors settings of PDF document, Fonts settings of PDF document. Simply drag and drop. Easily batch converts DjVu documents into PDFs from your documents. Feature Batch Converting DjVu documents into PDF normal files couldn't be any easier. Simply Drag the PUB files to the file list which you want to convert into PDF file and click the "Convert to PDF And Save as" button, A-PDF DjVu to PDF quickly re- creates your DjVu file as a fully formatted PDF files. -

Audio File Type Guide Digital Stewardship Curriculum Uncompressed Lossless
Audio File Type Guide Digital Stewardship Curriculum There are three types of audio quality: uncompressed, lossless, and lossy. Uncompressed audio keeps all the quality of the original source intact, lossless compresses audio for less space while maintaining quality, and lossy audio compresses the files more for saving space at a slightly reduced quality. To keep it simple, we recommend using WAV file format (uncompressed) for your archival master copy, and MP3 (lossy) as your access copy. However, here are explanations of these and several other audio formats in a bit more detail. Uncompressed ● WAV: An uncompressed format, which means it is an exact copies of the original source audio. The standard audio file format used mainly in Windows PCs. Commonly used for storing uncompressed (PCM), CD-quality sound files, which means that they can be large in size - around 10MB per minute of music. ● AIFF: AIFF is made by Apple, so you may see it a bit more often in Apple products. Also uncompressed, this format is like WAV, but a little less widely used. Since both these formats are uncompressed, they take up a lot of space. Lossless ● FLAC: The Free Lossless Audio Codec (FLAC) is the most popular lossless format, making it a good choice if you want to store your music in lossless. Unlike WAV and AIFF, it's been compressed, so it takes up a lot less space. However, it's still a lossless format, which means the audio quality is still the same as the original source, so it's much better for listening than WAV and AIFF. -

An Overview of Ogg Vorbis
An Overview of Ogg Vorbis Presented by: Shi Yong MUMT 611 Overview • Ogg – A multimedia container format, contains audio and video data streams in a single file, similar to mov and avi. – Functions: framing, sync, error correction, positioning – Stream oriented, suitable for internet streaming. – Used with various Codecs: • Vorbis: Audio codec • Tremor: Fixed-point decoder • Theora: Video codec • FLAC: Free Lossless Audio Codec • Speex: Speech codec • OggWrit: Text phrase codec (subtitles) • Ogg Metadata: Arbitrary metadata format Overview • Vorbis – General purpose lossy audio compression algorithm/format, similar to MP3, WMA, etc.. – Designed to be contained in a transport mechanism that provides framing, sync and error correction functions, such as Ogg (for file transport) or RTP (for webcast) – When used with Ogg, it is called Ogg Vorbis. – A great number of player now support Ogg Vorbis – Encode CD/DAT quality at below 48kbps – Wide range of sample rates: from 8kHz to 192kHz – Strong channel representations: monaural, polyphonic, stereo, quadraphonic, 5.1, up to 255 discrete channels Overview • Both Ogg and Vorbis are developed by the Xiph.Org Foundation, a non-profit corporation (http://www.xiph.org/ ) • LGPL licensing: – open standard – open source – patent free – completely free for commercial or noncommercial use • Software can be downloaded from http://www.vorbis.com/setup/ Encode • Overview – Accepting input audio, dividing it into frames, compressing frames into raw packets – As a generic perceptual audio encoder, psycho acoustic model is used to remove redundant audio information – Function blocks is similar to most other lossy audio encoders, such as Time/Frequency Analysis, Psychoacoustic Analysis, Quantization and Encoding, Bit Allocation, Entropy Coding, etc. -
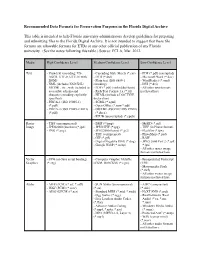
Recommended Data Formats for Preservation Purposes in the Florida Digital Archive
Recommended Data Formats for Preservation Purposes in the Florida Digital Archive This table is intended to help Florida university administrators develop guidelines for preparing and submitting files to the Florida Digital Archive. It is not intended to suggest that these file formats are allowable formats for ETDs or any other official publication of any Florida university. (See the notes following this table.) Source: FCLA, Mar. 2012. Media High Confidence Level Medium Confidence Level Low Confidence Level Text - Plain text (encoding: US- - Cascading Style Sheets (*.css) - PDF (*.pdf) (encrypted) ASCII, UTF-8, UTF-16 with - DTD (*.dtd) - Microsoft Word (*.doc) BOM) - Plain text (ISO 8859-1 - WordPerfect (*.wpd) - XML (includes XSD/XSL/ encoding) - DVI (*.dvi) XHTML, etc.; with included or - PDF (*.pdf) (embedded fonts) - All other text formats accessible schema and - Rich Text Format 1.x (*.rtf) not listed here character encoding explicitly - HTML (include a DOCTYPE specified) declaration) - PDF/A-1 (ISO 19005-1) - SGML (*.sgml) (*.pdf) - Open Office (*.sxw/*.odt) - PDF/A-2 (ISO 19005-2:2011) - OOXML (ISO/IEC DIS 29500) (*.pdf) (*.docx) - EPUB (unencrypted) (*.epub) Raster - TIFF (uncompressed) - BMP (*.bmp) - MrSID (*.sid) Image - JPEG2000 (lossless) (*.jp2) - JPEG/JFIF (*.jpg) - TIFF (in Planar format) - PNG (*.png) - JPEG2000 (lossy) (*.jp2) - FlashPix (*.fpx) - TIFF (compressed) - PhotoShop (*.psd) - GIF (*.gif) - RAW - Digital Negative DNG (*.dng) - JPEG 2000 Part 2 (*.jpf, - Google WebP (*.webp) *.jpx) - All other raster image -

FOSS Open Standards/Comparison of File Formats 1 FOSS Open Standards/Comparison of File Formats
FOSS Open Standards/Comparison of File Formats 1 FOSS Open Standards/Comparison of File Formats FOSS Open Standards Foreword — Preface — List of Acronyms — Introduction — Importance and Benefits of Open Standards — Standard-Setting and Open Standards — Some Important Open Standards — Comparison of File Formats — Standards and Internationalization/Localization of Software — Patents in Standards — The Linux Standard Base — Government/National Open Standards Policies and Initiatives — Conclusion — Annexure: Comments on RAND, as seen from both sides — Glossary — About the Author — Acknowledgements — About APDIP — About IOSN This section will list, compare and discuss the degrees of openness and/or lack of openness of several popular file formats. These include file formats for the following application areas: 1. office applications 2. graphics 3. audio 4. video Office Applications File Formats Microsoft Office Formats Currently, the most popular office application is Microsoft Office (MS Office). This suite of office software comprises mainly (depending on the type of suite purchased) word processing (MS Word), spreadsheet (MS Excel) and presentation software (MS PowerPoint). Up till version 10 (MS Office 10), the file formats used were binary (i.e. non-plain text) in nature and not publicly published. MS Word, MS Excel and MS PowerPoint use the binary DOC, XLS and PPT formats, respectively, and these are proprietary formats, being owned and controlled entirely by Microsoft. The file formats for these applications are widely used due to the popularity of MS Office. Other software not from Microsoft, e.g. OpenOffice.org or StarOffice, are able to read and write files using these proprietary formats but the compatibility is incomplete.