Influence of Pathogenic Bacterial Determinants on Genome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Endolysosomes and Inter-Organellar Signaling in Brain Disease
University of North Dakota UND Scholarly Commons Biomedical Sciences Faculty Publications Department of Biomedical Sciences 2-2020 Role of endolysosomes and inter-organellar signaling in brain disease Zahra Afghah Xuesong Chen University of North Dakota, [email protected] Jonathan David Geiger University of North Dakota, [email protected] Follow this and additional works at: https://commons.und.edu/bms-fac Part of the Medicine and Health Sciences Commons Recommended Citation Afghah, Zahra; Chen, Xuesong; and Geiger, Jonathan David, "Role of endolysosomes and inter-organellar signaling in brain disease" (2020). Biomedical Sciences Faculty Publications. 1. https://commons.und.edu/bms-fac/1 This Article is brought to you for free and open access by the Department of Biomedical Sciences at UND Scholarly Commons. It has been accepted for inclusion in Biomedical Sciences Faculty Publications by an authorized administrator of UND Scholarly Commons. For more information, please contact [email protected]. Neurobiology of Disease 134 (2020) 104670 Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi Review Role of endolysosomes and inter-organellar signaling in brain disease T ⁎ Zahra Afghah, Xuesong Chen, Jonathan D. Geiger Department of Biomedical Sciences, University of North Dakota School of Medicine and Health Sciences, Grand Forks, North Dakota 58201, United States of America ARTICLE INFO ABSTRACT Keywords: Endosomes and lysosomes (endolysosomes) are membrane bounded organelles that play a key role in cell sur- Endolysosomes vival and cell death. These acidic intracellular organelles are the principal sites for intracellular hydrolytic Mitochondria activity required for the maintenance of cellular homeostasis. -
Participant List
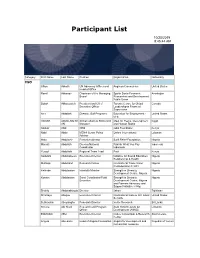
Participant List 10/20/2019 8:45:44 AM Category First Name Last Name Position Organization Nationality CSO Jillian Abballe UN Advocacy Officer and Anglican Communion United States Head of Office Ramil Abbasov Chariman of the Managing Spektr Socio-Economic Azerbaijan Board Researches and Development Public Union Babak Abbaszadeh President and Chief Toronto Centre for Global Canada Executive Officer Leadership in Financial Supervision Amr Abdallah Director, Gulf Programs Educaiton for Employment - United States EFE HAGAR ABDELRAHM African affairs & SDGs Unit Maat for Peace, Development Egypt AN Manager and Human Rights Abukar Abdi CEO Juba Foundation Kenya Nabil Abdo MENA Senior Policy Oxfam International Lebanon Advisor Mala Abdulaziz Executive director Swift Relief Foundation Nigeria Maryati Abdullah Director/National Publish What You Pay Indonesia Coordinator Indonesia Yussuf Abdullahi Regional Team Lead Pact Kenya Abdulahi Abdulraheem Executive Director Initiative for Sound Education Nigeria Relationship & Health Muttaqa Abdulra'uf Research Fellow International Trade Union Nigeria Confederation (ITUC) Kehinde Abdulsalam Interfaith Minister Strength in Diversity Nigeria Development Centre, Nigeria Kassim Abdulsalam Zonal Coordinator/Field Strength in Diversity Nigeria Executive Development Centre, Nigeria and Farmers Advocacy and Support Initiative in Nig Shahlo Abdunabizoda Director Jahon Tajikistan Shontaye Abegaz Executive Director International Insitute for Human United States Security Subhashini Abeysinghe Research Director Verite -

See Who Attended
Company Name First Name Last Name Job Title Country 24Sea Gert De Sitter Owner Belgium 2EN S.A. George Droukas Data analyst Greece 2EN S.A. Yannis Panourgias Managing Director Greece 3E Geert Palmers CEO Belgium 3E Baris Adiloglu Technical Manager Belgium 3E David Schillebeeckx Wind Analyst Belgium 3E Grégoire Leroy Product Manager Wind Resource Modelling Belgium 3E Rogelio Avendaño Reyes Regional Manager Belgium 3E Luc Dewilde Senior Business Developer Belgium 3E Luis Ferreira Wind Consultant Belgium 3E Grégory Ignace Senior Wind Consultant Belgium 3E Romain Willaime Sales Manager Belgium 3E Santiago Estrada Sales Team Manager Belgium 3E Thomas De Vylder Marketing & Communication Manager Belgium 4C Offshore Ltd. Tom Russell Press Coordinator United Kingdom 4C Offshore Ltd. Lauren Anderson United Kingdom 4Cast GmbH & Co. KG Horst Bidiak Senior Product Manager Germany 4Subsea Berit Scharff VP Offshore Wind Norway 8.2 Consulting AG Bruno Allain Président / CEO Germany 8.2 Consulting AG Antoine Ancelin Commercial employee Germany 8.2 Monitoring GmbH Bernd Hoering Managing Director Germany A Word About Wind Zoe Wicker Client Services Manager United Kingdom A Word About Wind Richard Heap Editor-in-Chief United Kingdom AAGES Antonio Esteban Garmendia Director - Business Development Spain ABB Sofia Sauvageot Global Account Executive France ABB Jesús Illana Account Manager Spain ABB Miguel Angel Sanchis Ferri Senior Product Manager Spain ABB Antoni Carrera Group Account Manager Spain ABB Luis andres Arismendi Gomez Segment Marketing Manager Spain -

A Foundation for the Future
A FOUNDATION FOR THE FUTURE INVESTORS REPORT 2012–13 NORTHWESTERN UNIVERSITY Dear alumni and friends, As much as this is an Investors Report, it is also living proof that a passion for collaboration continues to define the Kellogg community. Your collective support has powered the forward movement of our ambitious strategic plan, fueled development of our cutting-edge curriculum, enabled our global thought leadership, and helped us attract the highest caliber of students and faculty—all key to solidifying our reputation among the world’s elite business schools. This year, you also helped set a new record for alumni support of Kellogg. Our applications and admissions numbers are up dramatically. We have outpaced our peer schools in career placements for new graduates. And we have broken ground on our new global hub. Your unwavering commitment to everything that Kellogg stands for helps make all that possible. Your continuing support keeps us on our trajectory to transform business education and practice to meet the challenges of the new economy. Thank you for investing in Kellogg today and securing the future for generations of courageous leaders to come. All the best, Sally Blount ’92, Dean 4 KELLOGG.NORTHWESTERN.EDU/INVEST contentS 6 Transforming Together 8 Early Investors 10 Kellogg Leadership Circle 13 Kellogg Investors Leaders Partners Innovators Activators Catalysts who gave $1,000 to $2,499 who gave up to $1,000 99 Corporate Affiliates 101 Kellogg Investors by Class Year 1929 1949 1962 1975 1988 2001 1934 1950 1963 1976 1989 2002 -

Public Notices & the Courts
PUBLIC NOTICES B1 DAILY BUSINESS REVIEW WEDNESDAY, SEPTEMBER 29, 2021 dailybusinessreview.com & THE COURTS MIAMI-DADE PUBLIC NOTICES BUSINESS LEADS THE COURTS WEB SEARCH FORECLOSURE NOTICES: Notices of Action, NEW CASES FILED: US District Court, circuit court, EMERGENCY JUDGES: Listing of emergency judges Search our extensive database of public notices for Notices of Sale, Tax Deeds B5 family civil and probate cases B2 on duty at night and on weekends in civil, probate, FREE. Search for past, present and future notices in criminal, juvenile circuit and county courts. Also duty Miami-Dade, Broward and Palm Beach. SALES: Auto, warehouse items and other BUSINESS TAX RECEIPTS (OCCUPATIONAL Magistrate and Federal Court Judges B14 properties for sale B6 LICENSES): Names, addresses, phone numbers Simply visit: and type of business of those who have received CALENDARS: Suspensions in Miami-Dade, Broward, https://www.law.com/dailybusinessreview/public-notices/ FICTITIOUS NAMES: Notices of intent business licenses B2 and Palm Beach. Confirmation of judges’ daily motion to register B10 calendars in Miami-Dade B14 To search foreclosure sales by sale date visit: MARRIAGE LICENSES: Name, date of birth FAMILY MATTERS: Marriage dissolutions, adoptions, https://www.law.com/dailybusinessreview/foreclosures/ and city of those issued marriage licenses B3 DIRECTORIES: Addresses, telephone numbers, and termination of parental rights B7 names, and contact information for circuit and CREDIT INFORMATION: Liens filed against PROBATE NOTICES: Notices to Creditors, -

Surname Given Age Date Page Maiden Note Aageberg Alice F. 101 11-Feb D-1 See Also Article Feb. 12, P. C-5 Abegg Frances J. 64 9
Surname Given Age Date Page Maiden Note Aageberg Alice F. 101 11-Feb D-1 See also article Feb. 12, p. C-5 Abegg Frances J. 64 9-Jun D-1 Abel Delores Ruth 57 9-Sep B-6 Abel Ralph 31-Dec D-5 Abell Thelma 80 18-Dec A-11 See article, p. A-11 Abney Raymond 83 15-Dec D-1 See article, p. D-1 Acheson Helen G. 61 26-Apr E-2 Adamczyk Richard J. 48 5-Nov C-10 Adamovich Michael 77 10-Feb D-1 Adams Altha B. 91 12-Jan C-2 Adams Corinne, Deputy 79 12-Mar C-3 See also article March 13, p. C-1 Adams Frances D. 53 10-Nov D-5 Malloy Adams Leonard Eugene 58 2-Jun D-3 (Preacher) Adams Marie Mae 75 10-Mar C-1 Adams Robert F. 72 11-Jan D-1 Adams Susan L. 35 3-Mar C-6 Adams Thomas A. 12 Februray 2 A-6 Adams Thomas P. 38 19-Jan A-8 Addison James Grover "J. G." 69 3-Apr C-1 Adelsperger Edward H. 69 25-Mar D-1 Adelsperger Helen 71 24-Nov B-5 Adkins Kathleen 70 3-Feb B-7 Adler Sophie 74 4-Feb D-1 Adolph Bernard 60 2-Oct C-2 Adzima Rose 26-May D-5 Kubeck Agee Hardy 66 13-Jan A-1 See article, p. A-1 Agerter Tim D. 22 19-May D-1 See also article, p. A-2 Ahearn Mary 82 22-Apr C-3 Doolin Akhtar Julie Wayman 26 15-Apr C-5 Alaimo Bartole (Bart) 66 24-Oct B-2 Alaimo Damiana 86 20-Jan C-5 Alarcon Damiana 89 26-Jun C-1 Albert Florence E. -

Diplomatic List – Fall 2018
United States Department of State Diplomatic List Fall 2018 Preface This publication contains the names of the members of the diplomatic staffs of all bilateral missions and delegations (herein after “missions”) and their spouses. Members of the diplomatic staff are the members of the staff of the mission having diplomatic rank. These persons, with the exception of those identified by asterisks, enjoy full immunity under provisions of the Vienna Convention on Diplomatic Relations. Pertinent provisions of the Convention include the following: Article 29 The person of a diplomatic agent shall be inviolable. He shall not be liable to any form of arrest or detention. The receiving State shall treat him with due respect and shall take all appropriate steps to prevent any attack on his person, freedom, or dignity. Article 31 A diplomatic agent shall enjoy immunity from the criminal jurisdiction of the receiving State. He shall also enjoy immunity from its civil and administrative jurisdiction, except in the case of: (a) a real action relating to private immovable property situated in the territory of the receiving State, unless he holds it on behalf of the sending State for the purposes of the mission; (b) an action relating to succession in which the diplomatic agent is involved as an executor, administrator, heir or legatee as a private person and not on behalf of the sending State; (c) an action relating to any professional or commercial activity exercised by the diplomatic agent in the receiving State outside of his official functions. -- A diplomatic agent’s family members are entitled to the same immunities unless they are United States Nationals. -

FALL 2021 COURSE BULLETIN School of Visual Arts Division of Continuing Education Fall 2021
FALL 2021 COURSE BULLETIN School of Visual Arts Division of Continuing Education Fall 2021 2 The School of Visual Arts has been authorized by the Association, Inc., and as such meets the Education New York State Board of Regents (www.highered.nysed. Standards of the art therapy profession. gov) to confer the degree of Bachelor of Fine Arts on graduates of programs in Advertising; Animation; The School of Visual Arts does not discriminate on the Cartooning; Computer Art, Computer Animation and basis of gender, race, color, creed, disability, age, sexual Visual Effects; Design; Film; Fine Arts; Illustration; orientation, marital status, national origin or other legally Interior Design; Photography and Video; Visual and protected statuses. Critical Studies; and to confer the degree of Master of Arts on graduates of programs in Art Education; The College reserves the right to make changes from Curatorial Practice; Design Research, Writing and time to time affecting policies, fees, curricula and other Criticism; and to confer the degree of Master of Arts in matters announced in this or any other publication. Teaching on graduates of the program in Art Education; Statements in this and other publications do not and to confer the degree of Master of Fine Arts on grad- constitute a contract. uates of programs in Art Practice; Computer Arts; Design; Design for Social Innovation; Fine Arts; Volume XCVIII number 3, August 1, 2021 Illustration as Visual Essay; Interaction Design; Published by the Visual Arts Press, Ltd., © 2021 Photography, Video and Related Media; Products of Design; Social Documentary Film; Visual Narrative; and to confer the degree of Master of Professional Studies credits on graduates of programs in Art Therapy; Branding; Executive creative director: Anthony P. -

Jan 10 Pages FINAL.Qxp
SUPPLEMENT TO © 2010 Crain Communications Inc. All rights reserved. January 2010 Guide to Industry Executives Names and titles of administrators SPONSORED BY at car companies in the United States Aston Martin North America 9920 Irvine Center Drive, Irvine, CA 92618 949-379-3100 CEO VP & General Manager, Aston Martin North America Ulrich Bez Julian Jenkins AutoAlliance International Inc. 1 International Drive, Flat Rock, MI 48134 734-782-7800 Chairman President, CEO & Plant Manager VP Purchasing & Corporate Planning, CFO VP Human Resources General Manager, Operations & Launch Mazda General Manager Thomas Pixton John Savona Don Gelinas Rex Johnson Greg Brown Ikuo Sugiyama BMW of North America 300 Chestnut Ridge Road, Woodcliff Lake, NJ 07677 201-307-4000 Chairman & CEO* President Executive VP Operations Executive VP Finance & Administration VP Marketing Jim O'Donnell Jim O'Donnell Peter Miles Stefan Sengewald Jack Pitney VP Communications VP Mini VP Aftersales VP Engineering, U.S. VP Motorcycles Tom Kowaleski Jim McDowell Dan Creed Tom Baloga Pieter de Waal VP Legal Affairs VP Government Relations VP Government Affairs VP Eastern Region VP Western Region Howard Harris Craig Helsing Robert Nitto Wayne Orchowski Robert Frisch General Manager General Manager Manager, Electric Vehicle Operations VP Central Region VP Southern Region New Car Sales Center Development & Strategy, BMW Group Arturo Pineiro Gene Donnelly Richard Brekus George Baldwin Rich Steinberg *BMW (US) Holding Corp. Mini Rolls-Royce Motor Cars NA 201-307-4117 Manager Manager Corp. Communications General Manager General Manager General Manager General Manager General Manager Manager VP Mini Marketing Sales President Manager Sales Promotions Eastern Region Western Region Southern Region Aftersales Retail Operation Jim McDowell Trudy Hardy Steve Saward Paul Ferraiolo Karen Vonder Meulen Andy Thomas Chuck Blessed James Solomon Matthew Lynch Richard Hart Nicholas Brown BMW Manufacturing Co. -

Sunday, July 25, 2021 Sunday, July 25, 2021
Program of the Meeting CALENDAR OF EVENTS * Presenting Author Sunday, July 25, 2021 Sunday, July 25, 2021 Council Symposium SAM Multi-Disciplinary Scientific Symposium 10:30 am - 11:30 am 10:30 am - 11:30 am -TRACK 1 -TRACK 4 SU-A-TRACK 1 International Council Symposium: AAPM SU-A-TRACK 4 Solid State PET in Radiology and Global Engagement; Planning for the Future Radiation Oncology: from SiPM to AI in the Clinic Moderator: Ana Maria Marques da Silva, Pontifica Universidade Moderator: Jun Zhang, The Ohio State University, Columbus, OH Catolica do RS, Porto Alegre, RS 10:30 am J. Zhang, The Ohio State University: Technologies and AI 10:30 am S. Avery, University of Pennsylvania: Global Medical Features of SiPM PET Instrumentation Physics Education and Training 10:55 am S. Bowen, University of Washington, School of Medicine: 10:38 am S. Parker, Wake Forest Baptist Health High Point Medical Clinical Considerations of SiPM PET in Radiology and Center: Global Need Assessment Radiation Oncology 10:46 am J. Palta, Virginia Commonwealth University: Global 11:20 am Q&A & Panel Discussion Collaborations 10:54 am R. Jeraj, University of Wisconsin: Global Research and SAM Therapy Scientific Symposium Scientific Innovation 10:30 am - 12:30 pm 11:02 am G. Kim, University of California, San Diego: Global Data and Information Exchange -TRACK 5 11:10 am S. Wadi-Ramahi, University of Pittsburgh Medical Center: SU-AB-TRACK 5 Affordable Cancer Care for All Global Clinical Education and Training Moderator: Laurence Edward Court, University of Texas MD 11:18 am Q&A Anderson Cancer Center, Houston, TX 10:30 am Introduction Students and Trainees 10:33 am C. -

Surname Index, Vol. 41-50 (1980-1989)
Surname Index to Volumes 41-50 The Colorado Genealogist ©2013 Colorado Genealogical Society Surname Index for Volumes 41-50 Introduction This index was scanned from the annual surname indexes published each year in The Colorado Genealogist using a ScanSnap scanner which had OCR technology overlaid on the original images of the scanned pages. Every attempt has been made to obtain an accurate “reading” of the information. There may still be errors in the transfer. Since each year had its own index and there were different editors, some of the indexes have a surname for every name and some have only one surname and then list all first names under it. Be aware of this when searching. All of the original indexes used the spellings that appeared in the original quarterly publications, some of which may have contained spelling or typographical errors. A prudent searcher will try various spellings of the surnames being searched. Searching Hints •This is a searchable pdf document and uses the same searching tools you would find in your pdf reader, such as Adobe Acrobat Reader. •Begin by typing in a surname only. You may have to search on a page for the given name, like you would in a printed index. Index Volume 41 (1980) . The Colorado Genealogist INDEX TO VOLUME 41-1980 , Lucy Ann 183 ATCHISON, EuphemiaW. 37 BARKER, James 177 , Mollie Marie 183 ATKENSONE, James 37 BARKER, Joel 4 AEBEY, Russel P. 37 AUDREY, William 37 BARKER, Joshua 39 ABERNATHY, Isaac 37 AULSERBROOK, Alfred 37 BARNARD, George 164 ABMATHY, Essie 37 AUSC (?), George 37 BARNES, Clark 39 ADAMS, Charles 37 AUSTIN, Harvey E. -

Who's Who Legal: Competition 2021
Who’s Who Legal: Competition 2021 Competition ................................................................................... 1 State Aid ...................................................................................... 38 Plaintiff ........................................................................................ 41 Economists .................................................................................. 45 Future Leaders - Partners............................................................. 58 Competition Future Leaders - Non-Partners ................................. 68 Future Leaders - Economists ........................................................ 77 Competition Country Firm First Name Surname Argentina Allende & Brea Julian Peña Esteban Baker McKenzie Sociedad Civil Rópolo Pablo Beccar Varela Agustín Waisman Bomchil Marcelo den Toom Bouzat Rosenkrantz & Asociados Gabriel Bouzat Cabanellas Etchebarne Kelly Guillermo Cabanellas Estudio O'Farrell Agustín Siboldi Marval O'Farrell Mairal Miguel del Pino Marval O'Farrell Mairal Santiago Del Rio Pérez Alati Grondona Benites & Arntsen Luis Barry Tanoira Cassagne Abogados Bernardo Cassagne Australia Allen & Overy LLP Peter McDonald Allens Fiona Crosbie Allens Jacqueline Downes Allens Carolyn Oddie Ashurst Peter Armitage Banco Chambers Peter Brereton SC Banco Chambers Ruth C A Higgins SC Banco Chambers Cameron Moore SC Clayton Utz Michael Corrigan Clayton Utz Bruce Lloyd Clayton Utz Elizabeth Richmond Clayton Utz Kirsten Webb Clifford Chance Dave Poddar Corrs Chambers