Title Hybrid Origins of Citrus Varieties Inferred from DNA Marker Analysis
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid
Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid Diaphorina Liberibacter citri Plant Name asiaticus Citrus Huanglongbing Psyllid Aegle marmelos (L.) Corr. Serr.: bael, Bengal quince, golden apple, bela, milva X Aeglopsis chevalieri Swingle: Chevalier’s aeglopsis X X Afraegle gabonensis (Swingle) Engl.: Gabon powder-flask X Afraegle paniculata (Schum.) Engl.: Nigerian powder- flask X Atalantia missionis (Wall. ex Wight) Oliv.: see Pamburus missionis X X Atalantia monophylla (L.) Corr.: Indian atalantia X Balsamocitrus dawei Stapf: Uganda powder- flask X X Burkillanthus malaccensis (Ridl.) Swingle: Malay ghost-lime X Calodendrum capense Thunb.: Cape chestnut X × Citroncirus webberi J. Ingram & H. E. Moore: citrange X Citropsis gilletiana Swingle & M. Kellerman: Gillet’s cherry-orange X Citropsis schweinfurthii (Engl.) Swingle & Kellerm.: African cherry- orange X Citrus amblycarpa (Hassk.) Ochse: djerook leemo, djeruk-limau X Citrus aurantiifolia (Christm.) Swingle: lime, Key lime, Persian lime, lima, limón agrio, limón ceutí, lima mejicana, limero X X Citrus aurantium L.: sour orange, Seville orange, bigarde, marmalade orange, naranja agria, naranja amarga X Citrus depressa Hayata: shiikuwasha, shekwasha, sequasse X Citrus grandis (L.) Osbeck: see Citrus maxima X Citrus hassaku hort. ex Tanaka: hassaku orange X Citrus hystrix DC.: Mauritius papeda, Kaffir lime X X Citrus ichangensis Swingle: Ichang papeda X Citrus jambhiri Lushington: rough lemon, jambhiri-orange, limón rugoso, rugoso X X Citrus junos Sieb. ex Tanaka: xiang -

Characteristic Volatile Fingerprints and Odor Activity Values in Different
molecules Article Characteristic Volatile Fingerprints and Odor Activity Values in Different Citrus-Tea by HS-GC-IMS and HS-SPME-GC-MS Heting Qi 1,2,3 , Shenghua Ding 1,2,3, Zhaoping Pan 1,2,3, Xiang Li 1,2,3 and Fuhua Fu 1,2,3,* 1 Longping Branch Graduate School, Hunan University, Changsha 410125, China; [email protected] (H.Q.); [email protected] (S.D.); [email protected] (Z.P.); [email protected] (X.L.) 2 Provincial Key Laboratory for Fruits and Vegetables Storage Processing and Quality Safety, Agricultural Product Processing Institute, Hunan Academy of Agricultural Sciences, Changsha 410125, China 3 Hunan Province International Joint Lab on Fruits & Vegetables Processing, Quality and Safety, Changsha 410125, China * Correspondence: [email protected]; Tel.: +86-0731-82873369 Received: 24 November 2020; Accepted: 16 December 2020; Published: 19 December 2020 Abstract: Citrus tea is an emerging tea drink produced from tea and the pericarp of citrus, which consumers have increasingly favored due to its potential health effects and unique flavor. This study aimed to simultaneously combine the characteristic volatile fingerprints with the odor activity values (OAVs) of different citrus teas for the first time by headspace gas chromatography-ion mobility spectrometry (HS-GC-IMS) and headspace solid-phase microextraction-gas chromatography-mass spectrometry (HS-SPME-GC-MS). Results showed that the establishment of a citrus tea flavor fingerprint based on HS-GC-IMS data can provide an effective means for the rapid identification and traceability of different citrus varieties. Moreover, 68 volatile compounds (OAV > 1) were identified by HS-SPME-GC-MS, which reflected the contribution of aroma compounds to the characteristic flavor of samples. -

Varietal Update
November 17, 2016 FRESH NEWSLETTER FOR A NEW SEASON We are pleased to introduce an updated grower newsletter format that continues to provide seasonal varietal information, report on Sunkist sales and marketing activities while also highlighting consumer and industry trends that inform our sales and marketing strategies. Our staff will continue to share important regulatory information with growers separately from the newsletter to ensure timely news is communicated with the necessary urgency. Additionally, we encourage grower members to leverage the industry resources noted at the end of this newsletter for regular updates on important industry issues. As always, your feedback is important to us. Please e-mail questions, comments and suggestions about the newsletter to [email protected]. VARIETAL UPDATE ORANGES The Valencia orange crop has finished as the Navel orange season ramps up. The Navel crop is starting off with very good flavor and internal color. Focus is currently on sizes 88 and smaller as we wait for the season to progress with better overall color and more sizing options. Organic Navel oranges have also started shipping, with good availability and slightly larger sizing than the conventional crop; organic navels are currently at an 88/72/113 peak. The first packs of Cara Cara Navel oranges are expected the week of November 21/28, with initial sizing projected to be 88/72/113/56. Blood oranges are expected to start shipping the week of December 5. LEMONS Demand for lemons continues to grow, with bookings ahead of the past two years. Shipments are currently from districts 1 (D1), 2 (D2) and 3 (D3). -

Writing As Aesthetic in Modern and Contemporary Japanese-Language Literature
At the Intersection of Script and Literature: Writing as Aesthetic in Modern and Contemporary Japanese-language Literature Christopher J Lowy A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2021 Reading Committee: Edward Mack, Chair Davinder Bhowmik Zev Handel Jeffrey Todd Knight Program Authorized to Offer Degree: Asian Languages and Literature ©Copyright 2021 Christopher J Lowy University of Washington Abstract At the Intersection of Script and Literature: Writing as Aesthetic in Modern and Contemporary Japanese-language Literature Christopher J Lowy Chair of the Supervisory Committee: Edward Mack Department of Asian Languages and Literature This dissertation examines the dynamic relationship between written language and literary fiction in modern and contemporary Japanese-language literature. I analyze how script and narration come together to function as a site of expression, and how they connect to questions of visuality, textuality, and materiality. Informed by work from the field of textual humanities, my project brings together new philological approaches to visual aspects of text in literature written in the Japanese script. Because research in English on the visual textuality of Japanese-language literature is scant, my work serves as a fundamental first-step in creating a new area of critical interest by establishing key terms and a general theoretical framework from which to approach the topic. Chapter One establishes the scope of my project and the vocabulary necessary for an analysis of script relative to narrative content; Chapter Two looks at one author’s relationship with written language; and Chapters Three and Four apply the concepts explored in Chapter One to a variety of modern and contemporary literary texts where script plays a central role. -

Organic Acids in the Juice of Acid Lemon and Japanese Acid Citrus by Gas Chromatography
九州大学学術情報リポジトリ Kyushu University Institutional Repository Organic Acids in the Juice of Acid Lemon and Japanese Acid Citrus by Gas Chromatography Widodo, Soesiladi E. Fruit Science Laboratory, Faculty of Agriculture, Kyushu University Shiraishi, Mikio Fruit Science Laboratory, Faculty of Agriculture, Kyushu University Shiraishi, Shinichi Fruit Science Laboratory, Faculty of Agriculture, Kyushu University https://doi.org/10.5109/24091 出版情報:九州大学大学院農学研究院紀要. 40 (1/2), pp.39-44, 1995-12. 九州大学農学部 バージョン: 権利関係: ,J. Fat. Agr., Kyushu IJniv., 40 (l-a), 39-44 (1995) Organic Acids in the Juice of Acid Lemon and Japanese Acid Citrus by Gas Chromatography Soesiladi E. Widodo, Mikio Shiraishi and Shinichi Shiraishi Fruit. Science Laboratory, Faculty of Agriculture, Kyushu University, Fukuoka 81 l-23, Japan (RWC~i/‘~?C~ C/1/?1P 15, 199<5) Acetate, glycolate, butyratc, oxalate, malonate, succinate, furnaratp, glyoxylate, malate, tattarate, cis-aconitatc and citrate were detected in the juice of Hanayu (Ci7tnt.s /ttrr/c~jrr Hart. ex Shirai), Daidai (C:. tr/i,rnt/tGt /II Linn. var. Cynthifera Y. Tanaka), Kabosu (6’. .sp/~rc:r~oc~r ~IXI Hart,. cx Tanaka), ‘Lisbon lemon (C.limon Burm. f. Lisbon) and Yuzu (C:.,jrr/ros Sieb. ex Tanaka) with compositions and contcnt,s varied according t,o sampling years and species. Citrate and rnalat,e were predominant, accounting for more than 90% and 3-9% of the total detected acids, respect.ively. The other acids presented in tracts, accounting t.ol.ally for roughly less than 0.5%. INTRODUCTION A number of chromatographic methods have been employed for determining organic acids (OAs) in citrus extracts. -
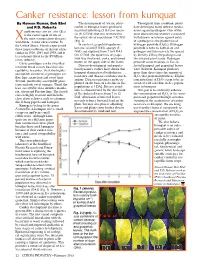
Canker Resistance: Lesson from Kumquat by Naveen Kumar, Bob Ebel the Development of Asiatic Citrus Throughout Their Evolution, Plants and P.D
Canker resistance: lesson from kumquat By Naveen Kumar, Bob Ebel The development of Asiatic citrus Throughout their evolution, plants and P.D. Roberts canker in kumquat leaves produced have developed many defense mecha- anthomonas citri pv. citri (Xcc) localized yellowing (5 DAI) or necro- nisms against pathogens. One of the is the causal agent of one of sis (9-12 DAI) that was restricted to most characteristic features associated the most serious citrus diseases the actual site of inoculation 7-12 DAI with disease resistance against entry X (Fig. 2). of a pathogen is the production of worldwide, Asiatic citrus canker. In the United States, Florida experienced In contrast, grapefruit epidermis hydrogen peroxide (H2O2). Hydrogen three major outbreaks of Asiatic citrus became raised (5 DAI), spongy (5 peroxide is toxic to both plant and canker in 1910, 1984 and 1995, and it DAI) and ruptured from 7 to 8 DAI. pathogen and thus restricts the spread is a constant threat to the $9 billion On 12 DAI, the epidermis of grape- by directly killing the pathogen and citrus industry. fruit was thickened, corky, and turned the infected plant tissue. Hydrogen Citrus genotypes can be classified brown on the upper side of the leaves. peroxide concentrations in Xcc-in- into four broad classes based on sus- Disease development and popula- fected kumquat and grapefruit leaves ceptibility to canker. First, the highly- tion dynamics studies have shown that were different. Kumquat produces susceptible commercial genotypes are kumquat demonstrated both disease more than three times the amount of Key lime, grapefruit and sweet lime. -

Freeze Response of Citrus and Citrus- Speeds (Nisbitt Et Al., 2000)
HORTSCIENCE 49(8):1010–1016. 2014. and tree and grove size (Bourgeois et al., 1990; Ebel et al., 2005). Protection using microsprinklers is compromised by high wind Freeze Response of Citrus and Citrus- speeds (Nisbitt et al., 2000). Developing more cold-tolerant citrus varieties through breeding related Genotypes in a Florida Field and selection has long been considered the most effective long-term solution (Grosser Planting et al., 2000; Yelenosky, 1985). Citrus and Citrus relatives are members Sharon Inch, Ed Stover1, and Randall Driggers of the family Rutaceae. The subtribe Citrinae U.S. Horticultural Research Laboratory, U.S. Department of Agriculture, is composed of Citrus (mandarins, oranges, Agricultural Research Service, 2001 South Rock Road, Fort Pierce, FL pummelos, grapefruits, papedas, limes, lem- ons, citrons, and sour oranges); Poncirus 34945 (deciduous trifoliate oranges); Fortunella Richard F. Lee (kumquats); Microcitrus and Eremocitrus (both Australian natives); and Clymenia National Clonal Germplasm Repository for Citrus and Dates, U.S. (Penjor et al., 2013). There is considerable Department of Agriculture, Agricultural Research Service, 1060 Martin morphological and ecological variation within Luther King Boulevard, Riverside, CA 92521 this group. With Citrus, cold-hardiness ranges from cold-tolerant to cold-sensitive (Soost and Additional index words. Aurantioideae, citrus breeding, cold-sensitive, defoliation, dieback, Roose, 1996). Poncirus and Fortunella are frost damage, Rutaceae, Toddalioideae considered the most cold-tolerant genera that Abstract. A test population consisting of progenies of 92 seed-source genotypes (hereafter are cross-compatible with Citrus. Poncirus called ‘‘parent genotypes’’) of Citrus and Citrus relatives in the field in east–central trifoliata reportedly can withstand tempera- Florida was assessed after natural freeze events in the winters of 2010 and 2011. -

Leading-Edge Technologies & Sophisticated Techniques Leading
Really The 2015-2016 Guidebook for the Database of Incredible! “SUGOWAZA” Manufacturing Companies in EHIME Database of“SUGOWAZA”Manufacturing Canpanies in EHIME Leading-edgeLeading-edge TechnologiesTechnologies& & SophisticatedSophisticated TechniquesTechniques, A Selection of 163 Companies! inEEHIMEHIME Leading-edge Technologies & Sophisticated Techniques in EEHIMEHIMAE Selection of 163 Companies! No.1 in Japan/ Only One is packed with information! There are many characteristic production companies accumulated in Ehime, varying in their regional history and culture. Information about the technology and products of these companies has been digitized in a database and publicized in this website. Complete with company search functions, and Ehime production industry introductions. Ehime Inquiries Prefecture ●Inquiries about "SUGOWAZA" database and registered companies Leading-edge Technologies & Sophisticated Techniques group, Industry Policy Division⦆Economy and Labor Department, Ehime Prefecture 4-4-2 Ichiban-cho, Matsuyama 790-8570 TEL: 089-912-2473 ⦆ FAX: 089-912-2259 E-mail:[email protected] http://www.sugowaza-ehime.com/ ehime sugowaza Search http://www.sugowaza-ehime.com/ 2015.07 m e s s a g e The industrial structure of Ehime Prefecture is defined by a balance rarely seen anywhere else in Japan, with each prefectural region having its own unique industrial concentrations: secondary industry is abundant in the Toyo Region (eastern area of the prefecture), tertiary industry thrives in the Chuyo Region (the central area of the prefecture around Total Value of Manufa ctured Goods Shipped Matsuyama City), and primary industry is dominant in the Nanyo Region (southwestern area of the prefecture). for Major Cities in Ehi me Prefecture Tokihiro Nakamura, There is a wide array of industrial cities in the Toyo Region, Governor of Ehime Prefecture which are home to numerous manufacturing companies boasting advanced technology unparalleled elsewhere in Japan and producing top-quality products. -
2019 Full Provisional List
Sheet1 All Plants Grafted. USDA inspected and Certified prior to Importing. Varieties Quantities Variety Description required Baboon Lemon A Brazilian lemon with very intense yellow rind and flesh. The flavour is acidic with almost a hint of lime. Tree is vigorous with large green leaves. Both tree and fruit are beautiful. Bearss Lemon 1952. Fruit closely resembles the Lisbon. Very juicy and has a high rind oil content. The leaves are a beautiful purple when first emerging, turning a nice dark green. Fruit is ready from June to December. Eureka Lemon Fruit is very juicy and highly acidic. The Eureka originated in Los Angeles, California and is one of their principal varieties. It is the "typical" lemon found in the grocery stores, nice yellow colour with typical lemon shape. Harvested November to May Harvey Lemon 1948.Having survived the disastrous deep freezes in Florida during the ’60’s and ’70’s. this varieties is known to withstand cold weather. Typical lemon shape and tart, juicy true lemon flavour. Fruit ripens in September to March. Self fertile. Zones 8A-10. Lisbon Lemon Fruit is very juicy and acic. The leaves are dense and tree is very vigorous. This Lisbon is more cold tolerant than the Eureka and is more productive. It is one of the major varieties in California. Fruit is harvested from February to May. Meyer Lemon 1908. Considered ever-bearing, the blooms are very aromatic. It is a lemon and orange hybrid. It is very cold hardy. Fruit is round with a thin rind. Fruit is juicy and has a very nice flavour, with a low acidity. -

The Confirmation of a Ploidy Periclinal Chimera of the Meiwa Kumquat
agronomy Article The Confirmation of a Ploidy Periclinal Chimera of the Meiwa Kumquat (Fortunella crassifolia Swingle) Induced by Colchicine Treatment to Nucellar Embryos and Its Morphological Characteristics 1, 1, 1, 1 1 Tsunaki Nukaya y, Miki Sudo y, Masaki Yahata *, Tomohiro Ohta , Akiyoshi Tominaga , Hiroo Mukai 1, Kiichi Yasuda 2 and Hisato Kunitake 3 1 Faculty of Agriculture, Shizuoka University, Shizuoka 422-8529, Japan; [email protected] (T.N.); [email protected] (M.S.); [email protected] (T.O.); [email protected] (A.T.); [email protected] (H.M.) 2 School of Agriculture, Tokai University, Kumamoto 862-8652, Japan; [email protected] 3 Faculty of Agriculture, University of Miyazaki, Miyazaki 889-2192, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-54-641-9500 These authors contributed equally to the article. y Received: 26 August 2019; Accepted: 12 September 2019; Published: 18 September 2019 Abstract: A ploidy chimera of the Meiwa kumquat (Fortunella crassifolia Swingle), which had been induced by treating the nucellar embryos with colchicine, and had diploid (2n = 2x = 18) and tetraploid (2n = 4x = 36) cells, was examined for its ploidy level, morphological characteristics, and sizes of its cells in its leaves, flowers, and fruits to reveal the ploidy level of each histogenic layer. Furthermore, the chimera was crossed with the diploid kumquat to evaluate the ploidy level of its reproductive organs. The morphological characteristics and the sizes of the cells in the leaves, flowers, and fruits of the chimera were similar to those of the tetraploid Meiwa kumquat and the ploidy periclinal chimera known as “Yubeni,” with diploids in the histogenic layer I (L1) and tetraploids in the histogenic layer II (L2) and III (L3). -

Citrus Fruits Information Compiled by Sunkist Growers
Commodity Fact Sheet Citrus Fruits Information compiled by Sunkist Growers How Produced – Citrus trees are propagated asexually mandarin orange and a lemon, are less acidic than traditional through a procedure known as grafting which fuses two lemons. different varieties of plants. In the case of citrus trees, one variety, the rootstock, is selected for Desert grapefruit are harvested October its hardiness and the other variety, the through March while summer grapefruit scion, is selected for its high-quality are available May through September. fruits. The rootstock, grown from a Specialty citrus include Melo Golds seed, is typically a two- to three-year- and Oro Blancos, grapefruit varieties old seedling while the scion is a bud that are popular with those preferring a from a mature tree. Through grafting, sweeter taste. Pummelos, or “Chinese” the scion fuses to the rootstock and grapefruit, considered a delicacy among becomes a new tree. In approximately many Asian cultures, are the largest of five years, the tree produces the same all citrus fruits. variety of fruit that was budded onto Almost a dozen different mandarin the rootstock. The successfully grafted and tangerine varieties, such as trees are sold to citrus growers through Clementines, Gold Nuggets, and wholesale nurseries and are certified Pixies, are available November through disease-free. There are approximately CITRUS VARIETIES May. Most are easy to peel and have a 270,000 bearing acres of citrus trees in lively flavor. California. Commodity Value – While Florida is the number one History – Oranges and lemons can be traced back to the producer of citrus fruits, the majority of their crop is made ancient Middle East. -
Holdings of the University of California Citrus Variety Collection 41
Holdings of the University of California Citrus Variety Collection Category Other identifiers CRC VI PI numbera Accession name or descriptionb numberc numberd Sourcee Datef 1. Citron and hybrid 0138-A Indian citron (ops) 539413 India 1912 0138-B Indian citron (ops) 539414 India 1912 0294 Ponderosa “lemon” (probable Citron ´ lemon hybrid) 409 539491 Fawcett’s #127, Florida collection 1914 0648 Orange-citron-hybrid 539238 Mr. Flippen, between Fullerton and Placentia CA 1915 0661 Indian sour citron (ops) (Zamburi) 31981 USDA, Chico Garden 1915 1795 Corsican citron 539415 W.T. Swingle, USDA 1924 2456 Citron or citron hybrid 539416 From CPB 1930 (Came in as Djerok which is Dutch word for “citrus” 2847 Yemen citron 105957 Bureau of Plant Introduction 3055 Bengal citron (ops) (citron hybrid?) 539417 Ed Pollock, NSW, Australia 1954 3174 Unnamed citron 230626 H. Chapot, Rabat, Morocco 1955 3190 Dabbe (ops) 539418 H. Chapot, Rabat, Morocco 1959 3241 Citrus megaloxycarpa (ops) (Bor-tenga) (hybrid) 539446 Fruit Research Station, Burnihat Assam, India 1957 3487 Kulu “lemon” (ops) 539207 A.G. Norman, Botanical Garden, Ann Arbor MI 1963 3518 Citron of Commerce (ops) 539419 John Carpenter, USDCS, Indio CA 1966 3519 Citron of Commerce (ops) 539420 John Carpenter, USDCS, Indio CA 1966 3520 Corsican citron (ops) 539421 John Carpenter, USDCS, Indio CA 1966 3521 Corsican citron (ops) 539422 John Carpenter, USDCS, Indio CA 1966 3522 Diamante citron (ops) 539423 John Carpenter, USDCS, Indio CA 1966 3523 Diamante citron (ops) 539424 John Carpenter, USDCS, Indio