Nova Proteins Direct Synaptic Integration of Somatostatin Interneurons Through Activity- Dependent Alternative Splicing
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-
![FK506-Binding Protein 12.6/1B, a Negative Regulator of [Ca2+], Rescues Memory and Restores Genomic Regulation in the Hippocampus of Aging Rats](https://docslib.b-cdn.net/cover/6136/fk506-binding-protein-12-6-1b-a-negative-regulator-of-ca2-rescues-memory-and-restores-genomic-regulation-in-the-hippocampus-of-aging-rats-16136.webp)
FK506-Binding Protein 12.6/1B, a Negative Regulator of [Ca2+], Rescues Memory and Restores Genomic Regulation in the Hippocampus of Aging Rats
This Accepted Manuscript has not been copyedited and formatted. The final version may differ from this version. A link to any extended data will be provided when the final version is posted online. Research Articles: Neurobiology of Disease FK506-Binding Protein 12.6/1b, a negative regulator of [Ca2+], rescues memory and restores genomic regulation in the hippocampus of aging rats John C. Gant1, Eric M. Blalock1, Kuey-Chu Chen1, Inga Kadish2, Olivier Thibault1, Nada M. Porter1 and Philip W. Landfield1 1Department of Pharmacology & Nutritional Sciences, University of Kentucky, Lexington, KY 40536 2Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294 DOI: 10.1523/JNEUROSCI.2234-17.2017 Received: 7 August 2017 Revised: 10 October 2017 Accepted: 24 November 2017 Published: 18 December 2017 Author contributions: J.C.G. and P.W.L. designed research; J.C.G., E.M.B., K.-c.C., and I.K. performed research; J.C.G., E.M.B., K.-c.C., I.K., and P.W.L. analyzed data; J.C.G., E.M.B., O.T., N.M.P., and P.W.L. wrote the paper. Conflict of Interest: The authors declare no competing financial interests. NIH grants AG004542, AG033649, AG052050, AG037868 and McAlpine Foundation for Neuroscience Research Corresponding author: Philip W. Landfield, [email protected], Department of Pharmacology & Nutritional Sciences, University of Kentucky, 800 Rose Street, UKMC MS 307, Lexington, KY 40536 Cite as: J. Neurosci ; 10.1523/JNEUROSCI.2234-17.2017 Alerts: Sign up at www.jneurosci.org/cgi/alerts to receive customized email alerts when the fully formatted version of this article is published. -

Genome-Wide Rnai Screening Identifies Human Proteins with A
RESOURCES Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway Jeremy C. Simpson1,7, Brigitte Joggerst2, Vibor Laketa2, Fatima Verissimo2, Cihan Cetin2, Holger Erfle2,6, Mariana G. Bexiga1, Vasanth R. Singan1, Jean-Karim Hériché3, Beate Neumann3, Alvaro Mateos2, Jonathon Blake4, Stephanie Bechtel5, Vladimir Benes4, Stefan Wiemann5, Jan Ellenberg2,3 and Rainer Pepperkok2,7 The secretory pathway in mammalian cells has evolved to facilitate the transfer of cargo molecules to internal and cell surface membranes. Use of automated microscopy-based genome-wide RNA interference screens in cultured human cells allowed us to identify 554 proteins influencing secretion. Cloning, fluorescent-tagging and subcellular localization analysis of 179 of these proteins revealed that more than two-thirds localize to either the cytoplasm or membranes of the secretory and endocytic pathways. The depletion of 143 of them resulted in perturbations in the organization of the COPII and/or COPI vesicular coat complexes of the early secretory pathway, or the morphology of the Golgi complex. Network analyses revealed a so far unappreciated link between early secretory pathway function, small GTP-binding protein regulation, actin cytoskeleton organization and EGF-receptor-mediated signalling. This work provides an important resource for an integrative understanding of global cellular organization and regulation of the secretory pathway in mammalian cells. Within higher eukaryotic cells membrane traffic pathways connect the Extensive efforts over many years have revealed a significant number various membrane-bounded organelles, thereby ensuring that they of regulators associated with the secretory pathway. Early biochemical retain the correct complement of proteins and lipids to maintain approaches to identify individual machinery components have started cellular homeostasis. -

Relevance Network Between Chemosensitivity and Transcriptome in Human Hepatoma Cells1
Vol. 2, 199–205, February 2003 Molecular Cancer Therapeutics 199 Relevance Network between Chemosensitivity and Transcriptome in Human Hepatoma Cells1 Masaru Moriyama,2 Yujin Hoshida, topoisomerase II  expression, whereas it negatively Motoyuki Otsuka, ShinIchiro Nishimura, Naoya Kato, correlated with expression of carboxypeptidases A3 Tadashi Goto, Hiroyoshi Taniguchi, and Z. Response to nimustine was associated with Yasushi Shiratori, Naohiko Seki, and Masao Omata expression of superoxide dismutase 2. Department of Gastroenterology, Graduate School of Medicine, Relevance networks identified several negative University of Tokyo, Tokyo 113-8655 [M. M., Y. H., M. O., N. K., T. G., H. T., Y. S., M. O.]; Cellular Informatics Team, Computational Biology correlations between gene expression and resistance, Research Center, Tokyo 135-0064 [S. N.]; and Department of which were missed by hierarchical clustering. Our Functional Genomics, Graduate School of Medicine, Chiba University, results suggested the necessity of systematically Chiba 260-8670 [N. S.], Japan evaluating the transporting systems that may play a major role in resistance in hepatoma. This may provide Abstract useful information to modify anticancer drug action in Generally, hepatoma is not a chemosensitive tumor, hepatoma. and the mechanism of resistance to anticancer drugs is not fully elucidated. We aimed to comprehensively Introduction evaluate the relationship between chemosensitivity and Hepatoma is a major cause of death even in developed gene expression profile in human hepatoma cells, by countries, and its incidence is increasing (1). Despite the using microarray analysis, and analyze the data by progress of therapeutic technique (2), the efficacy of radical constructing relevance networks. therapy is hampered by frequent recurrence and advance of In eight hepatoma cell lines (HLE, HLF, Huh7, Hep3B, the tumor (3). -

Genome-Wide Enrichment Analysis Between Endometriosis and Obesity-Related Traits Reveals Novel Susceptibility Loci
Human Molecular Genetics, 2015, Vol. 24, No. 4 1185–1199 doi:10.1093/hmg/ddu516 Advance Access published on October 8, 2014 Genome-wide enrichment analysis between endometriosis and obesity-related traits reveals novel susceptibility loci Nilufer Rahmioglu1, Stuart Macgregor2, Alexander W. Drong1,A˚ sa K. Hedman1,5, Holly R. Harris6,7, Joshua C. Randall8, Inga Prokopenko1,9,10, The International Endogene Consortium (IEC), The GIANT Consortium, Dale R. Nyholt3, Andrew P. Morris1,11,{, Grant W. Montgomery4,{, Downloaded from https://academic.oup.com/hmg/article/24/4/1185/617584 by guest on 05 February 2021 Stacey A. Missmer6,{, Cecilia M. Lindgren1,12,{ and Krina T. Zondervan1,13,∗,{ 1Wellcome Trust Center for Human Genetics, University of Oxford, Oxford OX3 7BN, UK, 2Statistical Genetics, 3Neurogenetics, 4Molecular Epidemiology, QIMR Berghofer Medical Research Institute, Brisbane, QLD 4029, Australia, 5Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden, 6Department of Obstetrics, Gynecology and Reproductive Biology, Brigham and Women’s Hospital and Harvard Medical School, 75 Francis Street, Boston, MA 02115, USA, 7Unit of Nutritional Epidemiology, Institute for Environmental Medicine, Karolinska Institutet, PO Box 210, SE-171 77 Stockholm, Sweden, 8Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK, 9Department of Genomics of Common Disease, Imperial College London, London W12 0NN, UK, 10Oxford Centre for Diabetes, Endocrinology and Metabolism, University -

The Abundance of Cis-Acting Loci Leading to Differential Allele
Yeo et al. BMC Genomics (2016) 17:620 DOI 10.1186/s12864-016-2922-9 RESEARCH ARTICLE Open Access The abundance of cis-acting loci leading to differential allele expression in F1 mice and their relationship to loci harboring genes affecting complex traits Seungeun Yeo1, Colin A. Hodgkinson1, Zhifeng Zhou1, Jeesun Jung2, Ming Leung1, Qiaoping Yuan1 and David Goldman1* Abstract Background: Genome-wide surveys have detected cis-acting quantitative trait loci altering levels of RNA transcripts (RNA-eQTLs) by associating SNV alleles to transcript levels. However, the sensitivity and specificity of detection of cis- expression quantitative trait loci (eQTLs) by genetic approaches, reliant as it is on measurements of transcript levels in recombinant inbred strains or offspring from arranged crosses, is unknown, as is their relationship to QTL’s for complex phenotypes. Results: We used transcriptome-wide differential allele expression (DAE) to detect cis-eQTLs in forebrain and kidney from reciprocal crosses between three mouse inbred strains, 129S1/SvlmJ, DBA/2J, and CAST/EiJ and C57BL/6 J. Two of these crosses were previously characterized for cis-eQTLs and QTLs for various complex phenotypes by genetic analysis of recombinant inbred (RI) strains. 5.4 %, 1.9 % and 1.5 % of genes assayed in forebrain of B6/ 129SF1, B6/DBAF1, and B6/CASTF1 mice, respectively, showed differential allelic expression, indicative of cis-acting alleles at these genes. Moreover, the majority of DAE QTLs were observed to be tissue-specific with only a small fraction showing cis-effects in both tissues. Comparing DAE QTLs in F1 mice to cis-eQTLs previously mapped in RI strains we observed that many of the cis-eQTLs were not confirmed by DAE. -

LHCGR (Luteinizing Hormone/Choriogonadotropin Receptor); LPHN2 (Latrophilin 2) (1P31.1)
Atlas of Genetics and Cytogenetics in Oncology and Haematology Home Genes Leukemias Solid Tumours Cancer-Prone Deep Insight Portal Teaching X Y 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 NA Atlas Journal Atlas Journal versus Atlas Database: the accumulation of the issues of the Journal constitutes the body of the Database/Text-Book. TABLE OF CONTENTS Volume 8, Number 1, Jan-Mar 2004 Previous Issue / Next Issue Genes ATBF1 (AT-binding transcription factor 1) (16q22.3-q23.1). Nadine Van Roy, Frank Speleman. Atlas Genet Cytogenet Oncol Haematol 2004; 8 (1): 1-7. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/ATBF1ID357.html DIRC2 (3q21). Anita Bonné, Danièlle Bodmer, Marc Eleveld, Eric Schoenmakers, Ad Geurts van Kessel.. Atlas Genet Cytogenet Oncol Haematol 2004; 8 (1): 8-11. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/DIRC2ID497.html DIRC3 (2q35). Anita Bonné, Danièlle Bodmer, Marc Eleveld, Eric Schoenmakers, Ad Geurts van Kessel.. Atlas Genet Cytogenet Oncol Haematol 2004; 8 (1): 12-14. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/DIRC3ID498..html RAP1GDS1 (4q22.3). Franck Viguié. Atlas Genet Cytogenet Oncol Haematol 2004; 8 (1): 15-18. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/RAP1GDS1ID400.html RET (REarranged during Transfection) (10q11.2). Patricia Niccoli-Sire. Atlas Genet Cytogenet Oncol Haematol 2004; 8 (1): 19-25. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/RETID76.html LHCGR (luteinizing hormone/choriogonadotropin receptor); LPHN2 (latrophilin 2) (1p31.1). Jim Heighway. Atlas Genet Cytogenet Oncol Haematol 2004; 8 (1): 26-36. -

A Post-Transcriptional Mechanism Pacing Expression of Neural Genes with Precursor Cell Differentiation Status
ARTICLE Received 24 Sep 2014 | Accepted 21 May 2015 | Published 6 Jul 2015 DOI: 10.1038/ncomms8576 OPEN A post-transcriptional mechanism pacing expression of neural genes with precursor cell differentiation status Weijun Dai1, Wencheng Li2, Mainul Hoque2, Zhuyun Li1, Bin Tian2 & Eugene V. Makeyev1,3 Nervous system (NS) development relies on coherent upregulation of extensive sets of genes in a precise spatiotemporal manner. How such transcriptome-wide effects are orchestrated at the molecular level remains an open question. Here we show that 30-untranslated regions (30 UTRs) of multiple neural transcripts contain AU-rich cis-elements (AREs) recognized by tristetraprolin (TTP/Zfp36), an RNA-binding protein previously implicated in regulation of mRNA stability. We further demonstrate that the efficiency of ARE-dependent mRNA degradation declines in the neural lineage because of a decrease in the TTP protein expression mediated by the NS-enriched microRNA miR-9. Importantly, TTP downregulation in this context is essential for proper neuronal differentiation. On the other hand, inactivation of TTP in non-neuronal cells leads to dramatic upregulation of multiple NS-specific genes. We conclude that the newly identified miR-9/TTP circuitry limits unscheduled accumulation of neuronal mRNAs in non-neuronal cells and ensures coordinated upregulation of these transcripts in neurons. 1 School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore. 2 Department of Microbiology, Biochemistry, and Molecular Genetics, Rutgers New Jersey Medical School, Newark, New Jersey 07103, USA. 3 MRC Centre for Developmental Neurobiology, King’s College London, London SE1 1UL, UK. Correspondence and requests for materials should be addressed to E.V.M. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

A Functional Enrichment Test for Molecular Convergent Evolution Finds a Clear Protein-Coding Signal in Echolocating Bats and Whales
A functional enrichment test for molecular convergent evolution finds a clear protein-coding signal in echolocating bats and whales Amir Marcovitza,1, Yatish Turakhiab,1, Heidi I. Chena,1, Michael Gloudemansc, Benjamin A. Braund, Haoqing Wange, and Gill Bejeranoa,d,f,g,2 aDepartment of Developmental Biology, Stanford University, Stanford, CA 94305; bDepartment of Electrical Engineering, Stanford University, Stanford, CA 94305; cBiomedical Informatics Program, Stanford University, Stanford, CA 94305; dDepartment of Computer Science, Stanford University, Stanford, CA 94305; eDepartment of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305; fDepartment of Pediatrics, Stanford University, Stanford, CA 94305; and gDepartment of Biomedical Data Science, Stanford University, Stanford, CA 94305 Edited by Scott V. Edwards, Harvard University, Cambridge, MA, and approved September 3, 2019 (received for review November 2, 2018) Distantly related species entering similar biological niches often parallel shifts in evolutionary rates in independent lineages of adapt by evolving similar morphological and physiological char- aquatic mammals (15). However, later analyses demonstrated that acters. How much genomic molecular convergence (particularly of the genome-wide frequency of molecular convergence in echolocating highly constrained coding sequence) contributes to convergent mammals is similar to the frequency in nonecholocating control phenotypic evolution, such as echolocation in bats and whales, is a outgroups, -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

The Human Gene Connectome As a Map of Short Cuts for Morbid Allele Discovery
The human gene connectome as a map of short cuts for morbid allele discovery Yuval Itana,1, Shen-Ying Zhanga,b, Guillaume Vogta,b, Avinash Abhyankara, Melina Hermana, Patrick Nitschkec, Dror Friedd, Lluis Quintana-Murcie, Laurent Abela,b, and Jean-Laurent Casanovaa,b,f aSt. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY 10065; bLaboratory of Human Genetics of Infectious Diseases, Necker Branch, Paris Descartes University, Institut National de la Santé et de la Recherche Médicale U980, Necker Medical School, 75015 Paris, France; cPlateforme Bioinformatique, Université Paris Descartes, 75116 Paris, France; dDepartment of Computer Science, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel; eUnit of Human Evolutionary Genetics, Centre National de la Recherche Scientifique, Unité de Recherche Associée 3012, Institut Pasteur, F-75015 Paris, France; and fPediatric Immunology-Hematology Unit, Necker Hospital for Sick Children, 75015 Paris, France Edited* by Bruce Beutler, University of Texas Southwestern Medical Center, Dallas, TX, and approved February 15, 2013 (received for review October 19, 2012) High-throughput genomic data reveal thousands of gene variants to detect a single mutated gene, with the other polymorphic genes per patient, and it is often difficult to determine which of these being of less interest. This goes some way to explaining why, variants underlies disease in a given individual. However, at the despite the abundance of NGS data, the discovery of disease- population level, there may be some degree of phenotypic homo- causing alleles from such data remains somewhat limited. geneity, with alterations of specific physiological pathways under- We developed the human gene connectome (HGC) to over- come this problem. -
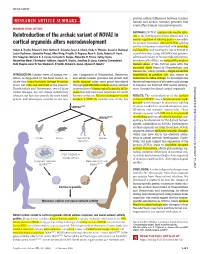
Reintroduction of the Archaic Variant of NOVA1
RESEARCH ◥ protein-coding differences between modern RESEARCH ARTICLE SUMMARY human and archaic hominin genomes that could affect human neurodevelopment. HUMAN EVOLUTION RATIONALE: NOVA1 regulates alternative splic- Reintroduction of the archaic variant of NOVA1 in ing in the developing nervous system and is a master regulator of splicing genes responsible cortical organoids alters neurodevelopment for synapse formation. Altered NOVA1 splicing activity in humans is associated with neurolog- Cleber A. Trujillo, Edward S. Rice, Nathan K. Schaefer, Isaac A. Chaim, Emily C. Wheeler, Assael A. Madrigal, ical disorders, underscoring the role of NOVA1 in Justin Buchanan, Sebastian Preissl, Allen Wang, Priscilla D. Negraes, Ryan A. Szeto, Roberto H. Herai, neural function. Using CRISPR-Cas9 genome- Alik Huseynov, Mariana S. A. Ferraz, Fernando S. Borges, Alexandre H. Kihara, Ashley Byrne, editing technology in human induced pluripo- Maximillian Marin, Christopher Vollmers, Angela N. Brooks, Jonathan D. Lautz, Katerina Semendeferi, tent stem cells (iPSCs), we replaced the modern Beth Shapiro, Gene W. Yeo, Stephen E. P. Smith, Richard E. Green, Alysson R. Muotri* human allele of the NOVA1 gene with the ancestral allele found in Neanderthals and Denisovans, which contains a single-nucleotide INTRODUCTION: Current views of human evo- cies. Comparison of Neanderthal, Denisovan, substitution at position 200 that causes an lution, as supported by the fossil record, in- and extant human genomes has shown that isoleucine-to-valine change. To investigate the dicate that many hominin lineage branches many humans today carry genes introduced functional importance of this amino acid change arose, but only one survived to the present. through past admixture events and has allowed in humans, we followed iPSC neural develop- Downloaded from Neanderthals and Denisovans, two of these enumeration of human-specific genetic differ- ment through functional cortical organoids.