Indene Bioconversion by a Toluene Inducible Dioxygenase of Rhodococcus Sp. I24
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

(12) United States Patent (10) Patent No.: US 8,323,956 B2 Reardon Et Al
USOO8323956B2 (12) United States Patent (10) Patent No.: US 8,323,956 B2 Reardon et al. (45) Date of Patent: Dec. 4, 2012 (54) DISTAL TIP OF BIOSENSORTRANSDUCER 6,159,681 A 12/2000 Zebala COMPRISINGENZYME FOR DEAMINATION 6,265,201 B1 7/2001 Wackett et al. 6,271,015 B1* 8/2001 Gilula et al. .................. 435,228 6,284.522 B1 9, 2001 Wackett et al. (75) Inventors: Kenneth F. Reardon, Fort Collins, CO 6,291,200 B1 9, 2001 LeJeune et al. (US); Lawrence Philip Wackett, St. 6,369,299 B1 4/2002 Sadowsky et al. 6.825,001 B2 11/2004 Wackett et al. Paul, MN (US) 7,381,538 B2 * 6/2008 Reardon et al. ................. 435/23 7.595,181 B2 * 9/2009 Gruning et al. ............... 435/197 (73) Assignee: Colorado State University Research 7,709,221 B2 5, 2010 Rose et al. Foundation, Fort Collins, CO (US) 2004/0265811 A1 12/2004 Reardon et al. 2006/0275855 A1 12/2006 Blackburn et al. .............. 435/15 (*) Notice: Subject to any disclaimer, the term of this FOREIGN PATENT DOCUMENTS patent is extended or adjusted under 35 U.S.C. 154(b) by 340 days. WO WOO3,O25892 12/1993 OTHER PUBLICATIONS (21) Appl. No.: 12/358,140 PCT/US2009/040121, International Search Report & Written Opin (22) Filed: Jan. 22, 2009 ion mailed Jul. 14, 2009, 8 pages. Adachi, K., et al; Purification and properties of homogentisate (65) Prior Publication Data oxygenase from Pseudomonas fluorescens. Biochim. Biophys. Acta 118 (1966) 88-97. US 2009/0221014 A1 Sep. 3, 2009 Aldridge, W.N.; Serum esterases. -

Indene CH Activation, Indole Π Vs. Nitrogen Lone-Pair Coordinati
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Organometallics Manuscript Author . Author Manuscript Author manuscript; available in PMC 2016 April 15. Published in final edited form as: Organometallics. 2007 ; 26(2): 281–287. doi:10.1021/om0606643. Reactions of Indene and Indoles with Platinum Methyl Cations: Indene C-H Activation, Indole π vs. Nitrogen Lone-Pair Coordination Travis J. Williams, Jay A. Labinger, and John E. Bercaw Arnold and Mabel Beckman Laboratories of Chemical Synthesis, California Institute of Technology, Pasadena, CA 91125 (U. S. A.) Abstract Reactions of indene and various substituted indoles with [(diimine)PtII(Me)(TFE)]+ cations have been studied (diimine = ArN = C(Me) − C(Me) = NAr; TFE = 2,2,2-trifluoroethanol). Indene displaces the TFE ligand from platinum to form a stable π coordination complex that, upon heating, undergoes C-H activation with first order kinetics, ΔH‡ = 29 kcal/mol, ΔS‡ = 10 eu, and a kinetic isotope effect of 1.1 at 60 °C. Indoles also initially form coordination complexes through the C2=C3 olefin, but these undergo rearrangement to the corresponding N-bound complexes. The relative rates of initial coordination and rearrangement are affected by excess acid or methyl substitution on indole. Introduction Selective C-H bond activation is a potentially valuable approach to synthetic problems in areas ranging from fuels and bulk chemicals to fine chemicals and pharmaceutical synthesis. 1 Studies of C-H activation in our laboratory have focused on models of the Shilov system,2 particularly [(diimine)PtII(Me)(solv)]+ (2, diimine = ArN = C(Me) − C(Me) = NAr; solv = 2,2,2-trifluoroethanol (TFE), H2O).3 These cations are capable of activating a variety of II carbon-hydrogen bonds.4 Cations 2 can be generated by protonolysis of (diimine)Pt Me2 species 1 in TFE with aqueous HBF43, 4ab or BX3 (X = C6F5,4cd F5), the latter producing H+ by boron coordination to TFE.4c In deuterated solvent 2 is formed as a mixture of two isotopologs. -

Thermophilic Bacteria Are Potential Sources of Novel Rieske Non-Heme
Chakraborty et al. AMB Expr (2017) 7:17 DOI 10.1186/s13568-016-0318-5 ORIGINAL ARTICLE Open Access Thermophilic bacteria are potential sources of novel Rieske non‑heme iron oxygenases Joydeep Chakraborty, Chiho Suzuki‑Minakuchi, Kazunori Okada and Hideaki Nojiri* Abstract Rieske non-heme iron oxygenases, which have a Rieske-type [2Fe–2S] cluster and a non-heme catalytic iron center, are an important family of oxidoreductases involved mainly in regio- and stereoselective transformation of a wide array of aromatic hydrocarbons. Though present in all domains of life, the most widely studied Rieske non-heme iron oxygenases are found in mesophilic bacteria. The present study explores the potential for isolating novel Rieske non- heme iron oxygenases from thermophilic sources. Browsing the entire bacterial genome database led to the identifi‑ cation of 45 homologs from thermophilic bacteria distributed mainly among Chloroflexi, Deinococcus–Thermus and Firmicutes. Thermostability, measured according to the aliphatic index, showed higher values for certain homologs compared with their mesophilic relatives. Prediction of substrate preferences indicated that a wide array of aromatic hydrocarbons could be transformed by most of the identified oxygenase homologs. Further identification of putative genes encoding components of a functional oxygenase system opens up the possibility of reconstituting functional thermophilic Rieske non-heme iron oxygenase systems with novel properties. Keywords: Rieske non-heme iron oxygenase, Oxidoreductase, Thermophiles, Aromatic hydrocarbons, Biotransformation Introduction of a wide array of agrochemically and pharmaceutically Rieske non-heme iron oxygenases (ROs) constitute a important compounds (Ensley et al. 1983; Wackett et al. large family of oxidoreductase enzymes involved primar- 1988; Hudlicky et al. -

Relating Metatranscriptomic Profiles to the Micropollutant
1 Relating Metatranscriptomic Profiles to the 2 Micropollutant Biotransformation Potential of 3 Complex Microbial Communities 4 5 Supporting Information 6 7 Stefan Achermann,1,2 Cresten B. Mansfeldt,1 Marcel Müller,1,3 David R. Johnson,1 Kathrin 8 Fenner*,1,2,4 9 1Eawag, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, 10 Switzerland. 2Institute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, 8092 11 Zürich, Switzerland. 3Institute of Atmospheric and Climate Science, ETH Zürich, 8092 12 Zürich, Switzerland. 4Department of Chemistry, University of Zürich, 8057 Zürich, 13 Switzerland. 14 *Corresponding author (email: [email protected] ) 15 S.A and C.B.M contributed equally to this work. 16 17 18 19 20 21 This supporting information (SI) is organized in 4 sections (S1-S4) with a total of 10 pages and 22 comprises 7 figures (Figure S1-S7) and 4 tables (Table S1-S4). 23 24 25 S1 26 S1 Data normalization 27 28 29 30 Figure S1. Relative fractions of gene transcripts originating from eukaryotes and bacteria. 31 32 33 Table S1. Relative standard deviation (RSD) for commonly used reference genes across all 34 samples (n=12). EC number mean fraction bacteria (%) RSD (%) RSD bacteria (%) RSD eukaryotes (%) 2.7.7.6 (RNAP) 80 16 6 nda 5.99.1.2 (DNA topoisomerase) 90 11 9 nda 5.99.1.3 (DNA gyrase) 92 16 10 nda 1.2.1.12 (GAPDH) 37 39 6 32 35 and indicates not determined. 36 37 38 39 S2 40 S2 Nitrile hydration 41 42 43 44 Figure S2: Pearson correlation coefficients r for rate constants of bromoxynil and acetamiprid with 45 gene transcripts of ECs describing nucleophilic reactions of water with nitriles. -

1/2 Toxic Compound Data Sheet Name: Indene CAS Number: 00095
1/2 Toxic Compound Data Sheet Name: Indene CAS Number: 00095-13-6 Justification: This compound is listed in Ohio Administrative Code 3745 - 114 - 01 because it fulfills one or more of the following criteria: substances that are known to be, or may reasonably be anticipated to be, carcinogenic, mutagenic, teratogenic, or neurotoxic, causes reproductive dysfunction, is acutely or chronically toxic, or causes the threat of adverse environmental effects through ambient concentrations, bioaccumulation, or atmospheric deposition. lndene is acutely toxic with effects on the upper respiratory system (mucous membranes), pulmonary irritation, liver and kidney effects. Molecular Weight (g/mol): 116.15 Synonyms: Indonaphthene U.S. EPA Carcinogenic Classification (IRIS): Not listed on IRIS. PBT: Not listed as persistent, bioaccumulative and toxic. NTP: Not listed by the National Toxicology Program. HAP: Not listed as a hazardous air pollutant (HAP) by U.S. EPA. 112r: Not listed under Section 112(r) of the Clean Air Act. ACGIH: TLV: 10 ppm or 47,505 µg/m3. Critical effects include upper respiratory irritation or damage, pulmonary irritation, and liver and kidney effects. HSDB: Listed in the Hazardous Substances Data Bank. Inhalation of indene vapors is expected to cause irritation of mucous membranes. International IARC: Not listed by International Agency for Research on Cancer (IARC). ATSDR (MRL): Not listed by ATSDR. DataSheet Indene.wpd 2/2 Reference Material 1. American Conference of Governmental Industrial Hygienists (ACGIH) 2006. TLVs and BEIs: -

Using SMILES Strings for the Description of Chemical Connectivity in the Crystallography Open Database
Quirós et al. J Cheminform (2018) 10:23 https://doi.org/10.1186/s13321-018-0279-6 RESEARCH ARTICLE Open Access Using SMILES strings for the description of chemical connectivity in the Crystallography Open Database Miguel Quirós1* , Saulius Gražulis2,3 , Saulė Girdzijauskaitė3, Andrius Merkys2 and Antanas Vaitkus2 Abstract Computer descriptions of chemical molecular connectivity are necessary for searching chemical databases and for predicting chemical properties from molecular structure. In this article, the ongoing work to describe the chemical connectivity of entries contained in the Crystallography Open Database (COD) in SMILES format is reported. This col- lection of SMILES is publicly available for chemical (substructure) search or for any other purpose on an open-access basis, as is the COD itself. The conventions that have been followed for the representation of compounds that do not ft into the valence bond theory are outlined for the most frequently found cases. The procedure for getting the SMILES out of the CIF fles starts with checking whether the atoms in the asymmetric unit are a chemically accept- able image of the compound. When they are not (molecule in a symmetry element, disorder, polymeric species,etc.), the previously published cif_molecule program is used to get such image in many cases. The program package Open Babel is then applied to get SMILES strings from the CIF fles (either those directly taken from the COD or those produced by cif_molecule when applicable). The results are then checked and/or fxed by a human editor, in a computer-aided task that at present still consumes a great deal of human time. -

Structure-Function Studies of Iron-Sulfur Enzyme Systems
Structure-Function Studies of Iron-Sulfur Enzyme Systems Rosmarie Friemann Department of Molecular Biology Uppsala Doctoral thesis Swedish University of Agricultural Sciences Uppsala 2005 1 Acta Universitatis Agriculturae Sueciae Agraria 504 ISSN 1401-6249 ISBN 91-576-6783-7 © 2004 Rosmarie Friemann, Uppsala Tryck: SLU Service/Repro, Uppsala 2004 2 Abstract Friemann, R., 2005, Structure-Function Studies of Iron-Sulfur Enzyme Systems. Doctorial dissertation. ISSN 1401-6249, ISBN 91-576-6783-7 Iron-sulfur clusters are among the most ancient of metallocofactors and serve a variety of biological functions in proteins, including electron transport, catalytic, and structural roles. Two kinds of multicomponent enzyme systems have been investigated by X-ray crystallography, the ferredoxin/thioredoxin system and bacterial Rieske non- heme iron dioxygenase (RDO) systems. The ferredoxin/thioredoxin system is a light sensitive system controlling the activities of key enzymes involved in the assimilatory (photosynthetic) and dissimilatory pathways in chloroplasts and photosynthetic bacteria. The system consists of a ferredoxin, ferredoxin:thioredoxin reductase (FTR), and two thioredoxins, Trx-m and Trx-f. In light, photosystem I reduces ferredoxin that reduces Trx-m and Trx- f. This two-electron reduction is catalyzed by FTR that contains a [4Fe-4S] center and a proximal disulfide bridge. When the first electron is delivered by the ferredoxin, an intermediate is formed where one thiol of the proximal disulfide attacks the disulfide bridge of thioredoxin. This results in a transient protein-protein complex held together by a mixed disulfide between FTR and Trx-m. This complex is stabilized by using a C40S mutant Trx-m and its structure have been determined. -
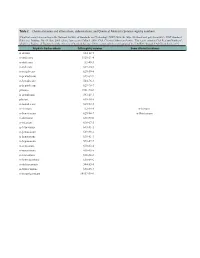
Table 2. Chemical Names and Alternatives, Abbreviations, and Chemical Abstracts Service Registry Numbers
Table 2. Chemical names and alternatives, abbreviations, and Chemical Abstracts Service registry numbers. [Final list compiled according to the National Institute of Standards and Technology (NIST) Web site (http://webbook.nist.gov/chemistry/); NIST Standard Reference Database No. 69, June 2005 release, last accessed May 9, 2008. CAS, Chemical Abstracts Service. This report contains CAS Registry Numbers®, which is a Registered Trademark of the American Chemical Society. CAS recommends the verification of the CASRNs through CAS Client ServicesSM] Aliphatic hydrocarbons CAS registry number Some alternative names n-decane 124-18-5 n-undecane 1120-21-4 n-dodecane 112-40-3 n-tridecane 629-50-5 n-tetradecane 629-59-4 n-pentadecane 629-62-9 n-hexadecane 544-76-3 n-heptadecane 629-78-7 pristane 1921-70-6 n-octadecane 593-45-3 phytane 638-36-8 n-nonadecane 629-92-5 n-eicosane 112-95-8 n-Icosane n-heneicosane 629-94-7 n-Henicosane n-docosane 629-97-0 n-tricosane 638-67-5 n-tetracosane 643-31-1 n-pentacosane 629-99-2 n-hexacosane 630-01-3 n-heptacosane 593-49-7 n-octacosane 630-02-4 n-nonacosane 630-03-5 n-triacontane 638-68-6 n-hentriacontane 630-04-6 n-dotriacontane 544-85-4 n-tritriacontane 630-05-7 n-tetratriacontane 14167-59-0 Table 2. Chemical names and alternatives, abbreviations, and Chemical Abstracts Service registry numbers.—Continued [Final list compiled according to the National Institute of Standards and Technology (NIST) Web site (http://webbook.nist.gov/chemistry/); NIST Standard Reference Database No. -

Biotechnological Potential of Rhodococcus Biodegradative Pathways Dockyu Kim1*, Ki Young Choi2, Miyoun Yoo3, Gerben J
J. Microbiol. Biotechnol. (2018), 28(7), 1037–1051 https://doi.org/10.4014/jmb.1712.12017 Research Article Review jmb Biotechnological Potential of Rhodococcus Biodegradative Pathways Dockyu Kim1*, Ki Young Choi2, Miyoun Yoo3, Gerben J. Zylstra4, and Eungbin Kim5 1Division of Polar Life Sciences, Korea Polar Research Institute, Incheon 21990, Republic of Korea 2University College, Yonsei University, Incheon 21983, Republic of Korea 3Korea Research Institute of Chemical Technology, Daejeon 34114, Republic of Korea 4Department of Biochemistry and Microbiology, School of Environmental and Biological Sciences, Rutgers University, NJ 08901-8520, USA 5Department of Systems Biology, Yonsei University, Seoul 03722, Republic of Korea Received: December 8, 2017 Revised: March 26, 2018 The genus Rhodococcus is a phylogenetically and catabolically diverse group that has been Accepted: May 1, 2018 isolated from diverse environments, including polar and alpine regions, for its versatile ability First published online to degrade a wide variety of natural and synthetic organic compounds. Their metabolic May 8, 2018 capacity and diversity result from their diverse catabolic genes, which are believed to be *Corresponding author obtained through frequent recombination events mediated by large catabolic plasmids. Many Phone: +82-32-760-5525; rhodococci have been used commercially for the biodegradation of environmental pollutants Fax: +82-32-760-5509; E-mail: [email protected] and for the biocatalytic production of high-value chemicals from low-value materials. -

Monohydroxylation of Phenoland 2,5-Dichlorophenol by Toluene
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, OCt. 1989, p. 2648-2652 Vol. 55, No. 10 0099-2240/89/102648-05$02.00/0 Copyright © 1989, American Society for Microbiology Monohydroxylation of Phenol and 2,5-Dichlorophenol by Toluene Dioxygenase in Pseudomonas putida Fl J. C. SPAIN,1* G. J. ZYLSTRA,2 C. K. BLAKE,2 AND D. T. GIBSON2 Air Force Engineering and Services Laboratory, Tyndall Air Force Base, Florida 32403,1 and Department of Microbiology, University of Iowa, Iowa City, Iowa 522422 Received 13 April 1989/Accepted 21 July 1989 Pseudomonas putida Fl contains a multicomponent enzyme system, toluene dioxygenase, that converts toluene and a variety of substituted benzenes to cis-dihydrodiols by the addition of one molecule of molecular oxygen. Toluene-grown cells of P. putida Fl also catalyze the monohydroxylation of phenols to the corresponding catechols by an unknown mechanism. Respirometric studies with washed cells revealed similar enzyme induction patterns in cells grown on toluene or phenol. Induction of toluene dioxygenase and subsequent enzymes for catechol oxidation allowed growth on phenol. Tests with specific mutants of P. putida Fl indicated that the ability to hydroxylate phenols was only expressed in cells that contained an active toluene dioxygenase enzyme system. 1802 experiments indicated that the overall reaction involved the incorporation of only one atom of oxygen in the catechol, which suggests either a monooxygenase mechanism or a dioxygenase reaction with subsequent specific elimination of water. The phenomenon of enzymatic oxygen fixation was first P. putida F39/D is a mutant strain of P. putida Fl that described in 1955 (11, 15). -

OSHA Permissible Exposure Limits (Pels) Are Too Permissive
Permissible Exposure Limits Copyright © 2018 Ronald N. Kostoff OSHA Permissible Exposure Limits (PELs) are too Permissive. by Dr. Ronald N. Kostoff Research Affiliate, School of Public Policy, Georgia Institute of Technology Gainesville, VA, 20155 Email: [email protected] KEYWORDS Permissible Exposure Limits; Exposure Limits; No-adverse-effects-exposure-limits; Minimal Risk Levels; NOAEL; OSHA; NIOSH; ACGIH; ATSDR 1 Permissible Exposure Limits Copyright © 2018 Ronald N. Kostoff CITATION TO MONOGRAPH Kostoff RN. OSHA Permissible Exposure Limits (PELs) are too Permissive. Georgia Institute of Technology. 2018. PDF. http://hdl.handle.net/1853/60067 COPYRIGHT AND CREATIVE COMMONS LICENSE COPYRIGHT Copyright © 2018 by Ronald N. Kostoff Printed in the United States of America; First Printing, 2018 CREATIVE COMMONS LICENSE This work can be copied and redistributed in any medium or format provided that credit is given to the original author. For more details on the CC BY license, see: http://creativecommons.org/licenses/by/4.0/ This work is licensed under a Creative Commons Attribution 4.0 International License<http://creativecommons.org/licenses/by/4.0/>. DISCLAIMERS The views in this monograph are solely those of the author, and do not represent the views of the Georgia Institute of Technology. 2 Permissible Exposure Limits Copyright © 2018 Ronald N. Kostoff TABLE OF CONTENTS TITLE KEYWORDS CITATION TO MONOGRAPH COPYRIGHT CREATIVE COMMONS LICENSE DISCLAIMERS ABSTRACT INTRODUCTION Background Relation of Biomedical Literature Findings to Setting of Exposure Limits Structure of Monograph ANALYSIS Overview Substance Selection Strategy Table 1 RESULTS AND DISCUSSION R-1. Acetaldehyde R-2. Trichloroethylene R-3. Naphthalene R-4. Dimethylbenzene R-5. Carbon Disulfide Interim Summary R-6. -

Carcinogens CAS DOT SHHC Sources Number Chemical Name
2010 Right to Know Special Health Hazardous Substance List Substance Common Name Carcinogens CAS DOT SHHC Sources Number Chemical Name 3140 # ACEPHATE 30560-19-1 2783 CA 3 6 8 17 18 PHOSPHORAMIDOTHIOIC ACID, ACETYL-, O,S-DIMETHYL ESTER 0001 # ACETALDEHYDE 75-07-0 1089 CA MU TE F4 1 2 3 4 5 6 7 8 R2 15 17 18 20 21 ACETALDEHYDE 22 2890 # ACETAMIDE 60-35-5 3077 CA 3 6 7 17 18 20 ACETAMIDE 0010 # 2-ACETYLAMINOFLUORENE 53-96-3 CA MU 1 4 5 6 18 20 21 ACETAMIDE, N-9H-FLUOREN-2-YL- 0022 # ACRYLAMIDE 79-06-1 2074 CA R2 1 2 3 4 5 6 7 8 15 17 18 19 20 2-PROPENAMIDE 21 0024 # ACRYLONITRILE 107-13-1 1093 CA TE F3 R2 1 2 3 4 5 6 7 8 14 15 17 18 19 2-PROPENENITRILE 20 21 22 3142 # AF- 2 3688-53-7 CA 7 2-FURANACETAMIDE, .alpha.-[(5-NITRO-2-FURANYL)METHYLENE]- 0029 # AFLATOXINS 1402-68-2 CA MU TE 5 7 AFLATOXINS 0033 # ALDRIN 309-00-2 2761 CA TE 1 2 3 4 6 7 8 14 17 18 19 20 21 1,4:5,8-DIMETHANONAPHTHALENE, 1,2,3,4,10,10-HEXACHLORO-1,4,4a,5,8,8aHEXAHYDRO(1R,4S,4aS,5S,8R,8aR)-rel- Page 1 of 55 2010 Right to Know Special Health Hazardous Substance List Substance Common Name Carcinogens CAS DOT SHHC Sources Number Chemical Name 0039 # ALLYL CHLORIDE 107-05-1 1100 CA F3 1 2 3 4 6 7 8 15 17 18 20 1-PROPENE, 3-CHLORO- 0069 # 2-AMINOANTHRAQUINONE 117-79-3 CA MU 5 6 7 18 9,10-ANTHRACENEDIONE, 2-AMINO- 4012 # 1-AMINO-2,4-DIBROMOANTHRAQUINONE 81-49-2 CA 5 9,10-ANTHRACENEDIONE, 1-AMINO-2,4-DIBROMO- 0072 # 4-AMINODIPHENYL 92-67-1 CA MU 1 2 4 5 6 7 18 20 [1,1'-BIPHENYL]-4-AMINE 0076 # 1-AMINO-2-METHYLANTHRAQUINONE 82-28-0 CA 5 6 7 18 9,10-ANTHRACENEDIONE, 1-AMINO-2-METHYL-