Supplementary Information Supplemental Materials
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Molecular Cytogenetic Analysis for TFE3 Rearrangement In
Modern Pathology (2014) 27, 113–127 & 2014 USCAP, Inc. All rights reserved 0893-3952/14 $32.00 113 Molecular cytogenetic analysis for TFE3 rearrangement in Xp11.2 renal cell carcinoma and alveolar soft part sarcoma: validation and clinical experience with 75 cases Jennelle C Hodge, Kathryn E Pearce, Xiaoke Wang, Anne E Wiktor, Andre M Oliveira and Patricia T Greipp Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA Renal cell carcinoma with TFE3 rearrangement at Xp11.2 is a distinct subtype manifesting an indolent clinical course in children, with recent reports suggesting a more aggressive entity in adults. This subtype is morphologically heterogeneous and can be misclassified as clear cell or papillary renal cell carcinoma. TFE3 is also rearranged in alveolar soft part sarcoma. To aid in diagnosis, a break-apart strategy fluorescence in situ hybridization (FISH) probe set specific for TFE3 rearrangement and a reflex dual-color, single-fusion strategy probe set involving the most common TFE3 partner gene, ASPSCR1, were validated on formalin-fixed, paraffin- embedded tissues from nine alveolar soft part sarcoma, two suspected Xp11.2 renal cell carcinoma, and nine tumors in the differential diagnosis. The impact of tissue cut artifact was reduced through inclusion of a chromosome X centromere control probe. Analysis of the UOK-109 renal carcinoma cell line confirmed the break-apart TFE3 probe set can distinguish the subtle TFE3/NONO fusion-associated inversion of chromosome X. Subsequent extensive clinical experience was gained through analysis of 75 cases with an indication of Xp11.2 renal cell carcinoma (n ¼ 54), alveolar soft part sarcoma (n ¼ 13), perivascular epithelioid cell neoplasms (n ¼ 2), chordoma (n ¼ 1), or unspecified (n ¼ 5). -

Rap-3971 Alveolar Soft Part Sarcoma Chromosome
DATA SHEET Alveolar Soft Part Sarcoma Chromosome Region, Candidate 1 Human Item Number rAP-3971 Synonyms ASPCR1, ASPL, ASPS, RCC17, TUG, UBXD9, UBXN9, Tether containing UBX domain for GLUT4, Alveo- lar soft part sarcoma chromosomal region candidate gene 1 protein, Alveolar soft part sarcoma locus, Re- nal papillary cell carcinoma protein 17, UBX domain-containin Description ASPSCR1 Human Recombinant produced in E. coli is a single polypeptide chain containing 576 amino acids (1-553) and having a molecular mass of 62.6kDa. ASPSCR1 is fused to a 23 amino acid His-tag at N- terminus & purified by proprietary chromatographic techniques. Uniprot Accesion Number Q9BZE9 Amino Acid Sequence MGSSHHHHHH SSGLVPRGSH MGSMAAPAGG GGSAVSVLAP NGRRHTVKVT PSTVLLQVLE DTCRRQDFNP CEYDLKFQRS VLDLSLQWRF ANLPNNAKLE MVPASRSREG PENMVRIALQ LDDGS- RLQDS FCSGQTLWEL LSHFPQIREC LQHPGGATPV CVYTRDEVTG EAALRGTTLQ SLGLTGGSAT IRFVMKCYDP VGKTPGSLGS SASAGQAAAS APLPLESGEL SRGDLSRPED ADTSGPCCEH TQEKQSTRAP AAAPFVPFSG GGQRLGGPPG PTRPLTSSSA KLPKSLSSPG GPSKPKKSKS GQDPQQEQEQ ERERDPQQEQ ERERPVDREP VDREPVVCHP DLEERLQAWP AELPDEFFEL Source Escherichia Coli. Physical Appearance Sterile Filtered clear solution. Store at 4°C if entire vial will be used within 2-4 weeks. Store, frozen at -20°C and Stability for longer periods of time. For long term storage it is recommended to add a carrier protein (0.1% HSA or BSA).Avoid multiple freeze-thaw cycles. Formulation and Purity The ASPSCR1 solution (0.25mg/ml) contains 20mM Tris-HCl buffer (pH 8.0), 0.15M NaCl, 10% glycerol and 1mM DTT. Greater than 85% as determined by SDS-PAGE. Application Solubility Biological Activity Shipping Format and Condition Lyophilized powder at room temperature. Optimal dilutions should be determined by each laboratory for each application. -

Chromosomal Aberrations in Head and Neck Squamous Cell Carcinomas in Norwegian and Sudanese Populations by Array Comparative Genomic Hybridization
825-843 12/9/08 15:31 Page 825 ONCOLOGY REPORTS 20: 825-843, 2008 825 Chromosomal aberrations in head and neck squamous cell carcinomas in Norwegian and Sudanese populations by array comparative genomic hybridization ERIC ROMAN1,2, LEONARDO A. MEZA-ZEPEDA3, STINE H. KRESSE3, OLA MYKLEBOST3,4, ENDRE N. VASSTRAND2 and SALAH O. IBRAHIM1,2 1Department of Biomedicine, Faculty of Medicine and Dentistry, University of Bergen, Jonas Lies vei 91; 2Department of Oral Sciences - Periodontology, Faculty of Medicine and Dentistry, University of Bergen, Årstadveien 17, 5009 Bergen; 3Department of Tumor Biology, Institute for Cancer Research, Rikshospitalet-Radiumhospitalet Medical Center, Montebello, 0310 Oslo; 4Department of Molecular Biosciences, University of Oslo, Blindernveien 31, 0371 Oslo, Norway Received January 30, 2008; Accepted April 29, 2008 DOI: 10.3892/or_00000080 Abstract. We used microarray-based comparative genomic logical parameters showed little correlation, suggesting an hybridization to explore genome-wide profiles of chromosomal occurrence of gains/losses regardless of ethnic differences and aberrations in 26 samples of head and neck cancers compared clinicopathological status between the patients from the two to their pair-wise normal controls. The samples were obtained countries. Our findings indicate the existence of common from Sudanese (n=11) and Norwegian (n=15) patients. The gene-specific amplifications/deletions in these tumors, findings were correlated with clinicopathological variables. regardless of the source of the samples or attributed We identified the amplification of 41 common chromosomal carcinogenic risk factors. regions (harboring 149 candidate genes) and the deletion of 22 (28 candidate genes). Predominant chromosomal alterations Introduction that were observed included high-level amplification at 1q21 (harboring the S100A gene family) and 11q22 (including Head and neck squamous cell carcinoma (HNSCC), including several MMP family members). -

The Endocytic Membrane Trafficking Pathway Plays a Major Role
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by University of Liverpool Repository RESEARCH ARTICLE The Endocytic Membrane Trafficking Pathway Plays a Major Role in the Risk of Parkinson’s Disease Sara Bandres-Ciga, PhD,1,2 Sara Saez-Atienzar, PhD,3 Luis Bonet-Ponce, PhD,4 Kimberley Billingsley, MSc,1,5,6 Dan Vitale, MSc,7 Cornelis Blauwendraat, PhD,1 Jesse Raphael Gibbs, PhD,7 Lasse Pihlstrøm, MD, PhD,8 Ziv Gan-Or, MD, PhD,9,10 The International Parkinson’s Disease Genomics Consortium (IPDGC), Mark R. Cookson, PhD,4 Mike A. Nalls, PhD,1,11 and Andrew B. Singleton, PhD1* 1Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, Maryland, USA 2Instituto de Investigación Biosanitaria de Granada (ibs.GRANADA), Granada, Spain 3Transgenics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, Maryland, USA 4Cell Biology and Gene Expression Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, Maryland, USA 5Department of Molecular and Clinical Pharmacology, Institute of Translational Medicine, University of Liverpool, Liverpool, United Kingdom 6Department of Pathophysiology, University of Tartu, Tartu, Estonia 7Computational Biology Group, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, Maryland, USA 8Department of Neurology, Oslo University Hospital, Oslo, Norway 9Department of Neurology and Neurosurgery, Department of Human Genetics, McGill University, Montréal, Quebec, Canada 10Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montréal, Quebec, Canada 11Data Tecnica International, Glen Echo, Maryland, USA ABSTRACT studies, summary-data based Mendelian randomization Background: PD is a complex polygenic disorder. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
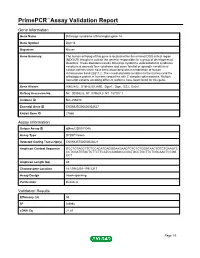
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DiGeorge syndrome critical region gene 14 Gene Symbol Dgcr14 Organism Mouse Gene Summary The human ortholog of this gene is located within the minimal DGS critical region (MDGCR) thought to contain the gene(s) responsible for a group of developmental disorders. These disorders include DiGeorge syndrome velocardiofacial syndrome conotruncal anomaly face syndrome and some familial or sporadic conotruncal cardiac defects which have been associated with microdeletion of human chromosome band 22q11.2. The encoded protein localizes to the nucleus and the orthologous protein in humans co-purifies with C complex spliceosomes. Multiple transcript variants encoding different isoforms have been found for this gene. Gene Aliases AI462402, D16H22S1269E, Dgcr1, Dgsi, ES2, Es2el RefSeq Accession No. NC_000082.6, NT_039624.8, NT_187007.1 UniGene ID Mm.256480 Ensembl Gene ID ENSMUSG00000003527 Entrez Gene ID 27886 Assay Information Unique Assay ID qMmuCID0011048 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000003621 Amplicon Context Sequence GCCTGTAGCTTCTCCACATCAGGGAAGAAGTCTCTCTGGATAACTGTCTGAAGTC CCTCGATGTACTCTTCTTCATCCAGGACCCGCTGCCTGCTTCTCGCAACTCCGG CCT Amplicon Length (bp) 82 Chromosome Location 16:17910201-17911217 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 95 R2 0.9994 cDNA Cq 21.87 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79.5 gDNA Cq 23.94 Specificity (%) 100 Information to assist with data interpretation is provided -

CRNKL1 Is a Highly Selective Regulator of Intron-Retaining HIV-1 and Cellular Mrnas
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 CRNKL1 is a highly selective regulator of intron-retaining HIV-1 and cellular mRNAs 2 3 4 Han Xiao1, Emanuel Wyler2#, Miha Milek2#, Bastian Grewe3, Philipp Kirchner4, Arif Ekici4, Ana Beatriz 5 Oliveira Villela Silva1, Doris Jungnickl1, Markus Landthaler2,5, Armin Ensser1, and Klaus Überla1* 6 7 1 Institute of Clinical and Molecular Virology, University Hospital Erlangen, Friedrich-Alexander 8 Universität Erlangen-Nürnberg, Erlangen, Germany 9 2 Berlin Institute for Medical Systems Biology, Max-Delbrück-Center for Molecular Medicine in the 10 Helmholtz Association, Robert-Rössle-Strasse 10, 13125, Berlin, Germany 11 3 Department of Molecular and Medical Virology, Ruhr-University, Bochum, Germany 12 4 Institute of Human Genetics, University Hospital Erlangen, Friedrich-Alexander Universität Erlangen- 13 Nürnberg, Erlangen, Germany 14 5 IRI Life Sciences, Institute für Biologie, Humboldt Universität zu Berlin, Philippstraße 13, 10115, Berlin, 15 Germany 16 # these two authors contributed equally 17 18 19 *Corresponding author: 20 Prof. Dr. Klaus Überla 21 Institute of Clinical and Molecular Virology, University Hospital Erlangen 22 Friedrich-Alexander Universität Erlangen-Nürnberg 23 Schlossgarten 4, 91054 Erlangen 24 Germany 25 Tel: (+49) 9131-8523563 26 e-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Mitochondrial Rab Gaps Govern Autophagosome Biogenesis During Mitophagy Koji Yamano1, Adam I Fogel1, Chunxin Wang1, Alexander M Van Der Bliek2, Richard J Youle1*
RESEARCH ARTICLE elife.elifesciences.org Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy Koji Yamano1, Adam I Fogel1, Chunxin Wang1, Alexander M van der Bliek2, Richard J Youle1* 1Biochemistry Section, Surgical Neurology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, United States; 2Department of Biological Chemistry, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, United States Abstract Damaged mitochondria can be selectively eliminated by mitophagy. Although two gene products mutated in Parkinson’s disease, PINK1, and Parkin have been found to play a central role in triggering mitophagy in mammals, how the pre-autophagosomal isolation membrane selectively and accurately engulfs damaged mitochondria remains unclear. In this study, we demonstrate that TBC1D15, a mitochondrial Rab GTPase-activating protein (Rab-GAP), governs autophagosome biogenesis and morphology downstream of Parkin activation. To constrain autophagosome morphogenesis to that of the cargo, TBC1D15 inhibits Rab7 activity and associates with both the mitochondria through binding Fis1 and the isolation membrane through the interactions with LC3/ GABARAP family members. Another TBC family member TBC1D17, also participates in mitophagy and forms homodimers and heterodimers with TBC1D15. These results demonstrate that TBC1D15 and TBC1D17 mediate proper autophagic encapsulation of mitochondria by regulating Rab7 activity at the interface between mitochondria and isolation membranes. DOI: 10.7554/eLife.01612.001 *For correspondence: [email protected] Introduction Competing interests: See page 21 Autophagosomes enclose seemingly random portions of the cytoplasm to supply nutrients during starvation or they can specifically engulf cellular debris to maintain quality control (Mizushima et al., Funding: See page 21 2011). -

1 UST College of Science Department of Biological Sciences
UST College of Science Department of Biological Sciences 1 Pharmacogenomics of Myofascial Pain Syndrome An Undergraduate Thesis Submitted to the Department of Biological Sciences College of Science University of Santo Tomas In Partial Fulfillment of the Requirements for the Degree of Bachelor of Science in Biology Jose Marie V. Lazaga Marc Llandro C. Fernandez May 2021 UST College of Science Department of Biological Sciences 2 PANEL APPROVAL SHEET This undergraduate research manuscript entitled: Pharmacogenomics of Myofascial Pain Syndrome prepared and submitted by Jose Marie V. Lazaga and Marc Llandro C. Fernandez, was checked and has complied with the revisions and suggestions requested by panel members after thorough evaluation. This final version of the manuscript is hereby approved and accepted for submission in partial fulfillment of the requirements for the degree of Bachelor of Science in Biology. Noted by: Asst. Prof. Marilyn G. Rimando, PhD Research adviser, Bio/MicroSem 602-603 Approved by: Bio/MicroSem 603 panel member Bio/MicroSem 603 panel member Date: Date: UST College of Science Department of Biological Sciences 3 DECLARATION OF ORIGINALITY We hereby affirm that this submission is our own work and that, to the best of our knowledge and belief, it contains no material previously published or written by another person nor material to which a substantial extent has been accepted for award of any other degree or diploma of a university or other institute of higher learning, except where due acknowledgement is made in the text. We also declare that the intellectual content of this undergraduate research is the product of our work, even though we may have received assistance from others on style, presentation, and language expression. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

A Multi-Stage Genome-Wide Association Study of Uterine Fibroids in African Americans
UCLA UCLA Previously Published Works Title A multi-stage genome-wide association study of uterine fibroids in African Americans. Permalink https://escholarship.org/uc/item/0mc5r0xh Journal Human genetics, 136(10) ISSN 0340-6717 Authors Hellwege, Jacklyn N Jeff, Janina M Wise, Lauren A et al. Publication Date 2017-10-01 DOI 10.1007/s00439-017-1836-1 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Hum Genet (2017) 136:1363–1373 DOI 10.1007/s00439-017-1836-1 ORIGINAL INVESTIGATION A multi‑stage genome‑wide association study of uterine fbroids in African Americans Jacklyn N. Hellwege1,2,3 · Janina M. Jef4 · Lauren A. Wise5,6 · C. Scott Gallagher7 · Melissa Wellons8,9 · Katherine E. Hartmann3,9 · Sarah F. Jones1,3 · Eric S. Torstenson1,2 · Scott Dickinson10 · Edward A. Ruiz‑Narváez6 · Nadin Rohland7 · Alexander Allen7 · David Reich7,11,12 · Arti Tandon7 · Bogdan Pasaniuc13,14 · Nicholas Mancuso13 · Hae Kyung Im10 · David A. Hinds15 · Julie R. Palmer6 · Lynn Rosenberg6 · Joshua C. Denny16,17 · Dan M. Roden2,16,17,18 · Elizabeth A. Stewart19 · Cynthia C. Morton12,20,21,22 · Eimear E. Kenny4 · Todd L. Edwards1,2,3 · Digna R. Velez Edwards2,3,9 Received: 12 April 2017 / Accepted: 16 August 2017 / Published online: 23 August 2017 © Springer-Verlag GmbH Germany 2017 Abstract Uterine fbroids are benign tumors of the uterus imaging, genotyped and imputed to 1000 Genomes. Stage 2 afecting up to 77% of women by menopause. They are the used self-reported fbroid and GWAS data from 23andMe, leading indication for hysterectomy, and account for $34 bil- Inc. -

The Orphan Disease Networks
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector ARTICLE The Orphan Disease Networks Minlu Zhang,1,3,5 Cheng Zhu,1,5 Alexis Jacomy,4 Long J. Lu,1,2,3 and Anil G. Jegga1,2,3,* The low prevalence rate of orphan diseases (OD) requires special combined efforts to improve diagnosis, prevention, and discovery of novel therapeutic strategies. To identify and investigate relationships based on shared genes or shared functional features, we have con- ducted a bioinformatic-based global analysis of all orphan diseases with known disease-causing mutant genes. Starting with a bipartite network of known OD and OD-causing mutant genes and using the human protein interactome, we first construct and topologically analyze three networks: the orphan disease network, the orphan disease-causing mutant gene network, and the orphan disease-causing mutant gene interactome. Our results demonstrate that in contrast to the common disease-causing mutant genes that are predomi- nantly nonessential, a majority of orphan disease-causing mutant genes are essential. In confirmation of this finding, we found that OD-causing mutant genes are topologically important in the protein interactome and are ubiquitously expressed. Additionally, func- tional enrichment analysis of those genes in which mutations cause ODs shows that a majority result in premature death or are lethal in the orthologous mouse gene knockout models. To address the limitations of traditional gene-based disease networks, we also construct and analyze OD networks on the basis of shared enriched features (biological processes, cellular components, pathways, phenotypes, and literature citations).