The Structure and Function of Large Biological Molecules 69 of Life
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Glossary - Cellbiology
1 Glossary - Cellbiology Blotting: (Blot Analysis) Widely used biochemical technique for detecting the presence of specific macromolecules (proteins, mRNAs, or DNA sequences) in a mixture. A sample first is separated on an agarose or polyacrylamide gel usually under denaturing conditions; the separated components are transferred (blotting) to a nitrocellulose sheet, which is exposed to a radiolabeled molecule that specifically binds to the macromolecule of interest, and then subjected to autoradiography. Northern B.: mRNAs are detected with a complementary DNA; Southern B.: DNA restriction fragments are detected with complementary nucleotide sequences; Western B.: Proteins are detected by specific antibodies. Cell: The fundamental unit of living organisms. Cells are bounded by a lipid-containing plasma membrane, containing the central nucleus, and the cytoplasm. Cells are generally capable of independent reproduction. More complex cells like Eukaryotes have various compartments (organelles) where special tasks essential for the survival of the cell take place. Cytoplasm: Viscous contents of a cell that are contained within the plasma membrane but, in eukaryotic cells, outside the nucleus. The part of the cytoplasm not contained in any organelle is called the Cytosol. Cytoskeleton: (Gk. ) Three dimensional network of fibrous elements, allowing precisely regulated movements of cell parts, transport organelles, and help to maintain a cell’s shape. • Actin filament: (Microfilaments) Ubiquitous eukaryotic cytoskeletal proteins (one end is attached to the cell-cortex) of two “twisted“ actin monomers; are important in the structural support and movement of cells. Each actin filament (F-actin) consists of two strands of globular subunits (G-Actin) wrapped around each other to form a polarized unit (high ionic cytoplasm lead to the formation of AF, whereas low ion-concentration disassembles AF). -

Oligomeric A2 + B3 Approach to Branched Poly(Arylene Ether Sulfone)
“One-Pot” Oligomeric A 2 + B 3 Approach to Branched Poly(arylene ether sulfone)s: Reactivity Ratio Controlled Polycondensation A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By ANDREA M. ELSEN B.S., Wright State University, 2007 2009 Wright State University WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES June 19, 200 9 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY Andrea M. Elsen ENTITLED “One-Pot” Oligomeric A2 + B3 Approach to Branched Poly(arylene ether sulfone)s: Reactivity Ratio Controlled Polycondenstation BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science . _________________________ Eric Fossum, Ph.D. Thesis Director _________________________ Kenneth Turnbull, Ph.D. Department Chair Committee on Final Examination ____________________________ Eric Fossum, Ph.D. ____________________________ Kenneth Turnbull, Ph.D. ____________________________ William A. Feld, Ph.D. ____________________________ Joseph F. Thomas, Jr., Ph.D. Dean, School of Graduate Studies Abstract Elsen, Andrea M. M.S., Department of Chemistry, Wright State University, 2009. “One-Pot” Oligomeric A 2 + B 3 Approach to Branched Poly(arylene ether sulfone)s: Reactivity Ratio Controlled Polycondensation The synthesis of fully soluble branched poly(arylene ether)s via an oligomeric A 2 + B 3 system, in which the A 2 oligomers are generated in situ, is presented. This approach takes advantage of the significantly higher reactivity toward nucleophilic aromatic substitution reactions, NAS, of B 2, 4-Fluorophenyl sulfone, relative to B 3, tris (4-Fluorophenyl) phosphine oxide. The A 2 oligomers were synthesized by reaction of Bisphenol-A and B 2, in the presence of the B 3 unit, at temperatures between 100 and 160 °C, followed by an increase in the reaction temperature to 180 °C at which point the branching unit was incorporated. -

Metabolic Engineering of Escherichia Coli for Poly(3-Hydroxybutyrate)
Lin et al. Microb Cell Fact (2015) 14:185 DOI 10.1186/s12934-015-0369-3 RESEARCH Open Access Metabolic engineering of Escherichia coli for poly(3‑hydroxybutyrate) production via threonine bypass Zhenquan Lin1,2,3†, Yan Zhang1,2,3†, Qianqian Yuan1,2,3,4, Qiaojie Liu1,2,3, Yifan Li1,2,3, Zhiwen Wang1,2,3, Hongwu Ma4*, Tao Chen1,2,3,5* and Xueming Zhao1,2,3 Abstract Background: Poly(3-hydroxybutyrate) (PHB), have been considered to be good candidates for completely biode- gradable polymers due to their similar mechanical properties to petroleum-derived polymers and complete biodeg- radability. Escherichia coli has been used to simulate the distribution of metabolic fluxes in recombinant E. coli pro- ducing poly(3-hydroxybutyrate) (PHB). Genome-scale metabolic network analysis can reveal unexpected metabolic engineering strategies to improve the production of biochemicals and biofuels. Results: In this study, we reported the discovery of a new pathway called threonine bypass by flux balance analysis of the genome-scale metabolic model of E. coli. This pathway, mainly containing the reactions for threonine synthesis and degradation, can potentially increase the yield of PHB and other acetyl-CoA derived products by reutilizing the CO2 released at the pyruvate dehydrogenase step. To implement the threonine bypass for PHB production in E. coli, we deregulated the threonine and serine degradation pathway and enhanced the threonine synthesis, resulting in 2.23-fold improvement of PHB titer. Then, we overexpressed glyA to enhance the conversion of glycine to serine and activated transhydrogenase to generate NADPH required in the threonine bypass. -
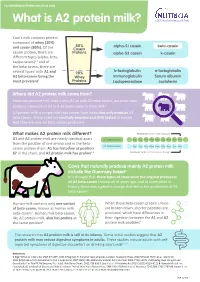
What Is A2 Protein Milk?
For Healthcare Professional use only What is A2 protein milk? Cow’s milk contains protein composed of whey (20%) 80% alpha-S1 casein beta-casein and casein (80%). Of the Casein casein protein, there are Proteins alpha-S2 casein k-casein different types (alpha, beta, kappa casein)1,2 and of the beta-casein, there are b-lactoglobulin a-lactoglobulin several types’ with A1 and 20% A2 beta-casein being the Whey Immunoglobulin Serum albumin most prevalent3 Proteins Lactoperoxidase Lactoferrin Where did A2 protein milk come from? Cows can produce milk that is only A1 or only A2 beta-casein, but most cows produce a mixture of A1 and A2 beta-casein in their milk4 A2 protein milk is simply milk that comes from cows that only produce A2 beta-casein. These cows are carefully selected and DNA tested to ensure that they are only A2 beta-casein producers What makes A2 protein milk different? Position 67(proline hinders cleavage) A1 and A2 protein milk are nearly identical apart A2 beta-casein Val Tyr Pro Phe Pro Gly Pro Iie Pro from the position of one amino acid in the beta- A1 beta-casein casein protein chain: A1 has histadine at position Val Tyr Pro Phe Pro Gly Pro Iie His 67 in the chain, and A2 protein milk has proline4,5 Position 67(histidine readily allows cleavage) Cows that naturally produce mainly A2 protein milk include the Guernsey breed4 It is thought that these types of cows were the original producers of A2 beta-casein thousands of years ago, and at some point in history there was a genetic change that led to the production of A1 beta-casein6 Human milk contains only one variant When these beta-casein protein chains of beta-casein, known as human milk are broken down, shorter peptides are beta-casein7. -

Protein Structure & Folding
6 Protein Structure & Folding To understand protein folding In the last chapter we learned that proteins are composed of amino acids Goal as a chemical equilibrium. linked together by peptide bonds. We also learned that the twenty amino acids display a wide range of chemical properties. In this chapter we will see Objectives that how a protein folds is determined by its amino acid sequence and that the three-dimensional shape of a folded protein determines its function by After this chapter, you should be able to: the way it positions these amino acids. Finally, we will see that proteins fold • describe the four levels of protein because doing so minimizes Gibbs free energy and that this minimization structure and the thermodynamic involves both making the most favorable bonds and maximizing disorder. forces that stabilize them. • explain how entropy (S) and enthalpy Proteins exhibit four levels of structure (H) contribute to Gibbs free energy. • use the equation ΔG = ΔH – TΔS The structure of proteins can be broken down into four levels of to determine the dependence of organization. The first is primary structure, the linear sequence of amino the favorability of a reaction on acids in the polypeptide chain. By convention, the primary sequence is temperature. written in order from the amino acid at the N-terminus (by convention • explain the hydrophobic effect and its usually on the left) to the amino acid at the C-terminus. The second level role in protein folding. of protein structure, secondary structure, is the local conformation adopted by stretches of contiguous amino acids. -

Casein Proteins As a Vehicle to Deliver Vitamin D3
Casein Proteins as a Vehicle to Deliver Vitamin D3: Fortification of Dairy Products with vitamin D3 and Bioavailability of Vitamin D3 from Fortified Mozzarella Cheese Baked with Pizza by Banaz Al-khalidi A thesis submitted in conformity with the requirements for the degree of Master of Science Graduate Department of Nutritional Sciences University of Toronto © Copyright by Banaz Al-khalidi (2012) Casein proteins as a vehicle to deliver vitamin D3: Fortification of dairy products with vitamin D3 and Bioavailability of vitamin D3 from fortified Mozzarella cheese baked with pizza Banaz Al-khalidi Master of Science Department of Nutritional Sciences, Faculty of Medicine University of Toronto 2012 ABSTRACT Current vitamin D intakes in Canada are inadequate. The extension of vitamin D fortification to additional foods may be an effective and appropriate strategy for increasing vitamin D intakes in the general population. Cheese is potentially an ideal candidate for vitamin D fortification. We introduce the potential use of casein proteins as a vehicle for vitamin D3 fortification in industrially made cheeses where we found that over 90% of vitamin D3 added to milk was retained in both Cheddar and Mozzarella cheeses. Use of casein proteins for vitamin D3 fortification did not fully prevent vitamin D3 loss into whey. However the loss was minimized to approximately 8%. We then show that vitamin D3 is bioavailable from fortified Mozzarella cheese baked with pizza suggesting that the high temperature baking process does not significantly breakdown vitamin D3. Our findings could have important implications in increasing fortified food options for Canadians. ii TABLE OF CONTENTS ABSTRACT .................................................................................................................................. -

DNA Glycosylase Exercise - Levels 1 & 2: Answer Key
Name________________________ StarBiochem DNA Glycosylase Exercise - Levels 1 & 2: Answer Key Background In this exercise, you will explore the structure of a DNA repair protein found in most species, including bacteria. DNA repair proteins move along DNA strands, checking for mistakes or damage. DNA glycosylases, a specific type of DNA repair protein, recognize DNA bases that have been chemically altered and remove them, leaving a site in the DNA without a base. Other proteins then come along to fill in the missing DNA base. Learning objectives We will explore the relationship between a protein’s structure and its function in a human DNA glycosylase called human 8-oxoguanine glycosylase (hOGG1). Getting started We will begin this exercise by exploring the structure of hOGG1 using a molecular 3-D viewer called StarBiochem. In this particular structure, the repair protein is bound to a segment of DNA that has been damaged. We will first focus on the structure hOGG1 and then on how this protein interacts with DNA to repair a damaged DNA base. • To begin using StarBiochem, please navigate to: http://mit.edu/star/biochem/. • Click on the Start button Click on the Start button for StarBiochem. • Click Trust when a prompt appears asking if you trust the certificate. • In the top menu, click on Samples à Select from Samples. Within the Amino Acid/Proteins à Protein tab, select “DNA glycosylase hOGG1 w/ DNA – H. sapiens (1EBM)”. “1EBM” is the four character unique ID for this structure. Take a moment to look at the structure from various angles by rotating and zooming on the structure. -

Quantitation of Relative Mobilities of Serum Transferrin in Quarter Horses and Thoroughbreds Using Crossed Immunoelectrophoresis
QUANTITATION OF RELATIVE MOBILITIES OF SERUM TRANSFERRIN IN QUARTER HORSES AND THOROUGHBREDS USING CROSSED IMMUNOELECTROPHORESIS BY RUTH GREER KAGEN II Bachelor of Science in Biology East Central Oklahoma State University Ada. Oklahoma 1982 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of requirements for the degree of MASTERS OF SCIENCE December. 1985 4 ' ;_ '" SERUM TRANSFERRIN IN QUARTER HORSES AND THOROUGHBREDS USING CROSSED IMMUNOELECTROPHORESIS Thesis Approved ii 1236431 ~ ACKNOWLEDGEMENTS The author wishes to express sincere gratitude to Dr. T. E. Staley for serving as the principle advisor and for providing the facilities and financial support necessary to carry out this work. I am most in debted to Dr. Staley for his continued faith in my work. his constant support and patient advice. and for his attentive and meticulous instruc tion as well as superb organization--all without which this study would not have been possible. I also wish to thank the members of the committee: Dr. C. L. Ownby and Dr. L. G. Stratton. I wish to extend my sincere thanks to Dr. C. L. Ownby for her continued interest in my career and her invaluable advice. both which greatly assisted me in the endeavors to carry out this work. I am very grateful for all the time which Dr. Ownby has afforded me and for her objective and sincere counsel. I am most appreciative of Dr. L. G. Stratton's interest in this project. Through his generous support and concern. we were provided with Pxcellent facilities in which to carry out this work and we were also provided with access to the animals needed for this project. -

Wilson Disease
Wilson Disease What is Wilson disease? Wilson disease is a hereditary disease in which excessive amounts of copper accumulate in the body, mainly in the liver. The disease affects approximately one in every 30,000 Canadians. Small amounts of copper are essential to good health. One of the liver’s jobs is to maintain the balance of copper in the body. The liver is also the main organ to store copper. In Wilson disease, when its storage capacity is full, copper is released into the blood stream. It then accumulates in various organs such as the brain and the cornea of the eye. This copper overload damages these organs. Left untreated, Wilson disease can be fatal. What causes the disease? Wilson disease is inherited. In order to have the disease, a person must get two defective genes, one from each parent. The liver begins to retain copper at birth and it may take years before symptoms manifest themselves. Having only one defective gene does not lead to Wilson disease. What are the symptoms of Wilson disease? Wilson disease is sometimes difficult to diagnose. Affected individuals may have no symptoms for years. When symptoms develop, they can be subtle. Sometimes symptoms of Wilson disease resemble hepatitis. Alternatively, some individuals have an enlarged liver and spleen and liver test abnormalities. Copper accumulation in the brain can present itself in two ways: (1) as physical symptoms such as slurred speech, failing voice, drooling, tremors or difficulty in swallowing or (2) as psychiatric disorders such as depression, manic behaviour or suicidal impulses. Very rarely, Wilson disease may cause the liver to fail. -

Chapter 23 Nucleic Acids
7-9/99 Neuman Chapter 23 Chapter 23 Nucleic Acids from Organic Chemistry by Robert C. Neuman, Jr. Professor of Chemistry, emeritus University of California, Riverside [email protected] <http://web.chem.ucsb.edu/~neuman/orgchembyneuman/> Chapter Outline of the Book ************************************************************************************** I. Foundations 1. Organic Molecules and Chemical Bonding 2. Alkanes and Cycloalkanes 3. Haloalkanes, Alcohols, Ethers, and Amines 4. Stereochemistry 5. Organic Spectrometry II. Reactions, Mechanisms, Multiple Bonds 6. Organic Reactions *(Not yet Posted) 7. Reactions of Haloalkanes, Alcohols, and Amines. Nucleophilic Substitution 8. Alkenes and Alkynes 9. Formation of Alkenes and Alkynes. Elimination Reactions 10. Alkenes and Alkynes. Addition Reactions 11. Free Radical Addition and Substitution Reactions III. Conjugation, Electronic Effects, Carbonyl Groups 12. Conjugated and Aromatic Molecules 13. Carbonyl Compounds. Ketones, Aldehydes, and Carboxylic Acids 14. Substituent Effects 15. Carbonyl Compounds. Esters, Amides, and Related Molecules IV. Carbonyl and Pericyclic Reactions and Mechanisms 16. Carbonyl Compounds. Addition and Substitution Reactions 17. Oxidation and Reduction Reactions 18. Reactions of Enolate Ions and Enols 19. Cyclization and Pericyclic Reactions *(Not yet Posted) V. Bioorganic Compounds 20. Carbohydrates 21. Lipids 22. Peptides, Proteins, and α−Amino Acids 23. Nucleic Acids ************************************************************************************** -

Gluten Free Casein Free Diet
GLUTEN FREE, CASEIN FREE DIET The gluten free, casein free (GFCF) diet has been shown to be helpful for individuals with allergies to these particular foods and specifically in the management of autistic spectrum disorder (ASD). Proteins found in grain and dairy products, known as gluten and casein respectively, are believed to be poorly broken down in the digestive tracts in some people. When these proteins are not digested properly they can be absorbed intact into blood circulation. These proteins can affect the brain by crossing the blood-brain barrier and binding to opioid receptors. This can affect mood, concentration, mental performance and pain tolerance (i.e. in autistic children this will increase their pain threshold). Research has shown significant improvement in several conditions, including schizophrenia and autism, following a GFCF diet. In a survey of over 3500 parents of autistic children, it was reported that 70% found a GFCF diet improved behavior, eye contact and socialisation, concentration and learning. It is recommended to follow the GFCF diet strictly for at least 6 months to assess the benefit of this diet. Below is a list of foods containing gluten and casein that are suggested to avoid, plus a list of alternative GFCF choices. RECOMMENDED AVOID GRAINS AND • Amaranth • Baked Beans unless gluten free LEGUMES • Basmati Rice • Flours: Wheat flour, wholemeal flour, • Beans bakers flour, semolina, barley, rye • Brown Rice (avoid battered or crumbed food) • Buckwheat • Wheat including durum, semolina, • Chickpea triticale, -

An O(N5) Algorithm for MFE Prediction of Kissing Hairpins and 4-Chains in Nucleic Acids
JOURNAL OF COMPUTATIONAL BIOLOGY Volume 16, Number 6, 2009 # Mary Ann Liebert, Inc. Pp. 803–815 DOI: 10.1089/cmb.2008.0219 An O(n5) Algorithm for MFE Prediction of Kissing Hairpins and 4-Chains in Nucleic Acids HO-LIN CHEN,1 ANNE CONDON,2 and HOSNA JABBARI2 ABSTRACT Efficient methods for prediction of minimum free energy (MFE) nucleic secondary struc- tures are widely used, both to better understand structure and function of biological RNAs and to design novel nano-structures. Here, we present a new algorithm for MFE secondary structure prediction, which significantly expands the class of structures that can be handled in O(n5) time. Our algorithm can handle H-type pseudoknotted structures, kissing hairpins, and chains of four overlapping stems, as well as nested substructures of these types. Key words: kissing hairpins, pseudoknot, RNA, secondary structure prediction. 1. INTRODUCTION ur knowledge of the amazing variety of functions played by RNA molecules in the cell Ocontinues to expand, with the functions determined in part by structure (Lee et al., 1997). Additionally, DNA and RNA sequences are designed to form novel structures for a wide range of applications, such as algorithmic DNA self-assembly (He et al., 2008; Rothemund et al., 2004), detection of low concentrations of other molecules of interest (Dirks and Pierce, 2004) or to exhibit motion (Simmel and Dittmer, 2005). In order to improve our ability to determine function from DNA or RNA sequences, and also to aid in the design of nucleic acids with novel structural or functional properties, accurate and efficient methods for predicting nucleic acid structure are very valuable.