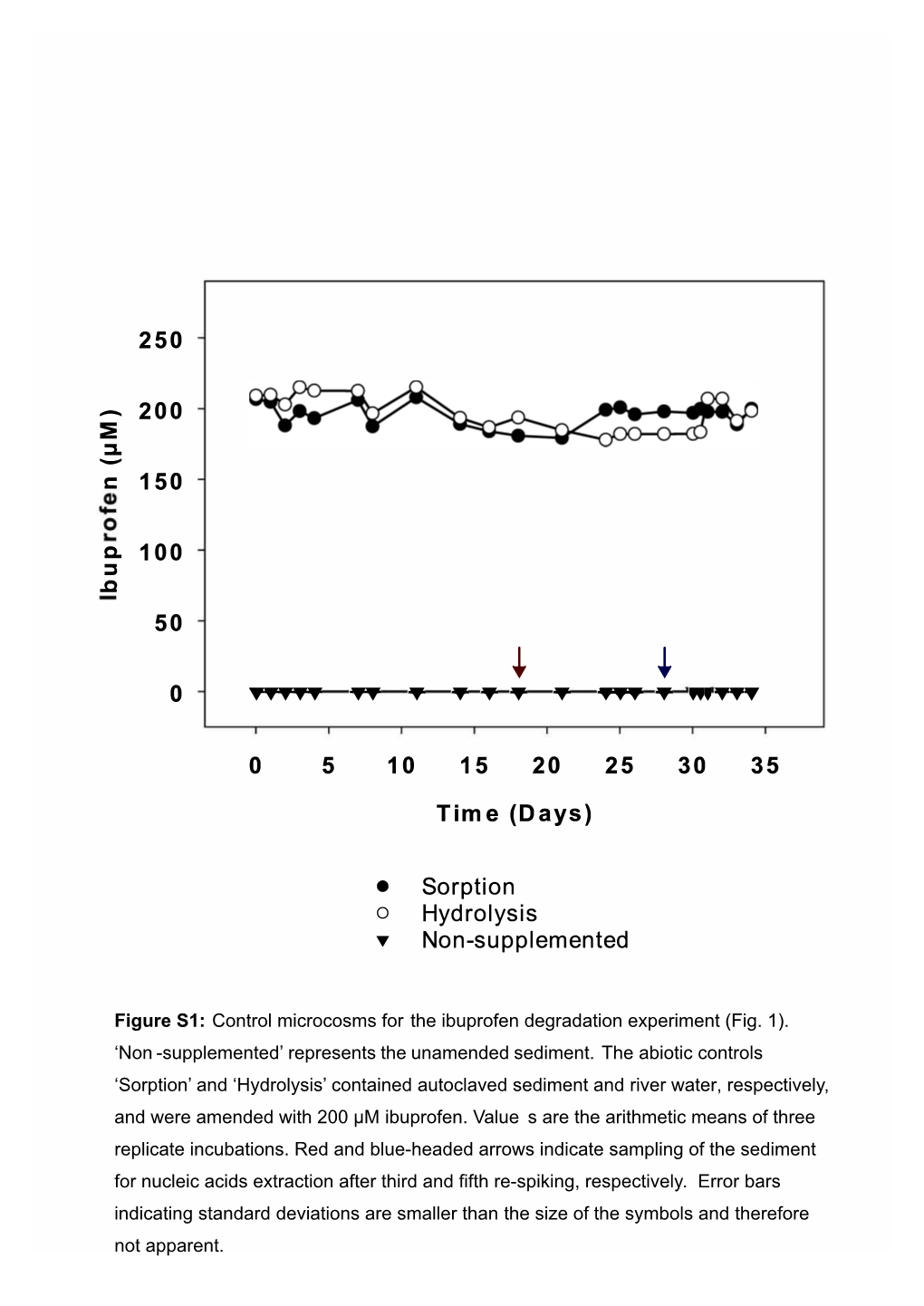
Non -Supplemented’ Represents the Unamended Sediment
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microbiology of Barrier Component Analogues of a Deep Geological Repository
Microbiology of Barrier Component Analogues of a Deep Geological Repository by Rachel Beaver A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Master of Science in Biology Waterloo, Ontario, Canada, 2020 ©Rachel Beaver 2020 Author’s Declaration This thesis consists of material all of which I authored or co-authored: see Statement of Contributions included in the thesis. This is a true copy of the thesis, including any required final revisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. ii Statement of Contributions Chapter 2 The Tsukinuno bentonite sampling was coordinated by Erik Kremmer (NWMO). The Opalinus core was received from Niels Burzan and Rizlan Bernier-Latmani (École Polytechnique Fédérale de Lausanne, Switzerland). The Northern Ontario crystalline rock core sampling was coordinated by Jeff Binns (Nuclear Waste Management Organization). Sian Ford (McMaster University) swabbed the outer layer of the Northern Ontario crystalline rocK core and crushed the inner layer. Melody Vachon (University of Waterloo) assisted with the cultivation of anaerobic heterotrophs and SRB. iii Abstract Many countries are in the process of designing and implementing long-term storage solutions for used nuclear fuel. Like many of these countries, Canada is considering a deep geological repository (DGR) wherein used fuel bundles will be placed in copper-coated carbon steel used fuel containers encased in bentonite buffer boxes. Previously published research has simulated aspects of a DGR experimentally to identify DGR conditions that would prevent microbial activity. Although such microcosm-type experiments can observe microbial growth and activity over relatively limited time frames, a DGR will remain functional for at least a million years, and will be exposed to fluctuating environmental conditions. -

The Role of Earthworm Gut-Associated Microorganisms in the Fate of Prions in Soil
THE ROLE OF EARTHWORM GUT-ASSOCIATED MICROORGANISMS IN THE FATE OF PRIONS IN SOIL Von der Fakultät für Lebenswissenschaften der Technischen Universität Carolo-Wilhelmina zu Braunschweig zur Erlangung des Grades eines Doktors der Naturwissenschaften (Dr. rer. nat.) genehmigte D i s s e r t a t i o n von Taras Jur’evič Nechitaylo aus Krasnodar, Russland 2 Acknowledgement I would like to thank Prof. Dr. Kenneth N. Timmis for his guidance in the work and help. I thank Peter N. Golyshin for patience and strong support on this way. Many thanks to my other colleagues, which also taught me and made the life in the lab and studies easy: Manuel Ferrer, Alex Neef, Angelika Arnscheidt, Olga Golyshina, Tanja Chernikova, Christoph Gertler, Agnes Waliczek, Britta Scheithauer, Julia Sabirova, Oleg Kotsurbenko, and other wonderful labmates. I am also grateful to Michail Yakimov and Vitor Martins dos Santos for useful discussions and suggestions. I am very obliged to my family: my parents and my brother, my parents on low and of course to my wife, which made all of their best to support me. 3 Summary.....................................................………………………………………………... 5 1. Introduction...........................................................................................................……... 7 Prion diseases: early hypotheses...………...………………..........…......…......……….. 7 The basics of the prion concept………………………………………………….……... 8 Putative prion dissemination pathways………………………………………….……... 10 Earthworms: a putative factor of the dissemination of TSE infectivity in soil?.………. 11 Objectives of the study…………………………………………………………………. 16 2. Materials and Methods.............................…......................................................……….. 17 2.1 Sampling and general experimental design..................................................………. 17 2.2 Fluorescence in situ Hybridization (FISH)………..……………………….………. 18 2.2.1 FISH with soil, intestine, and casts samples…………………………….……... 18 Isolation of cells from environmental samples…………………………….………. -

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Download Download
http://wjst.wu.ac.th Natural Sciences Diversity Analysis of an Extremely Acidic Soil in a Layer of Coal Mine Detected the Occurrence of Rare Actinobacteria Megga Ratnasari PIKOLI1,*, Irawan SUGORO2 and Suharti3 1Department of Biology, Faculty of Science and Technology, Universitas Islam Negeri Syarif Hidayatullah Jakarta, Ciputat, Tangerang Selatan, Indonesia 2Center for Application of Technology of Isotope and Radiation, Badan Tenaga Nuklir Nasional, Jakarta Selatan, Indonesia 3Department of Chemistry, Faculty of Science and Computation, Universitas Pertamina, Simprug, Jakarta Selatan, Indonesia (*Corresponding author’s e-mail: [email protected], [email protected]) Received: 7 September 2017, Revised: 11 September 2018, Accepted: 29 October 2018 Abstract Studies that explore the diversity of microorganisms in unusual (extreme) environments have become more common. Our research aims to predict the diversity of bacteria that inhabit an extreme environment, a coal mine’s soil with pH of 2.93. Soil samples were collected from the soil at a depth of 12 meters from the surface, which is a clay layer adjacent to a coal seam in Tanjung Enim, South Sumatera, Indonesia. A culture-independent method, the polymerase chain reaction based denaturing gradient gel electrophoresis, was used to amplify the 16S rRNA gene to detect the viable-but-unculturable bacteria. Results showed that some OTUs that have never been found in the coal environment and which have phylogenetic relationships to the rare actinobacteria Actinomadura, Actinoallomurus, Actinospica, Streptacidiphilus, Aciditerrimonas, and Ferrimicrobium. Accordingly, the highly acidic soil in the coal mine is a source of rare actinobacteria that can be explored further to obtain bioactive compounds for the benefit of biotechnology. -

Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens
microorganisms Article Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens Maryam Rezadehbashi and Susan A. Baldwin * Chemical and Biological Engineering, University of British Columbia, 2360 East Mall, Vancouver, BC V6T 1Z3, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-604-822-1973 Received: 2 January 2018; Accepted: 17 February 2018; Published: 23 February 2018 Abstract: Biochemical reactors (BCRs) based on the stimulation of sulphate-reducing microorganisms (SRM) are emerging semi-passive remediation technologies for treatment of mine-influenced water. Their successful removal of metals and sulphate has been proven at the pilot-scale, but little is known about the types of SRM that grow in these systems and whether they are diverse or restricted to particular phylogenetic or taxonomic groups. A phylogenetic study of four established pilot-scale BCRs on three different mine sites compared the diversity of SRM growing in them. The mine sites were geographically distant from each other, nevertheless the BCRs selected for similar SRM types. Clostridia SRM related to Desulfosporosinus spp. known to be tolerant to high concentrations of copper were members of the core microbial community. Members of the SRM family Desulfobacteraceae were dominant, particularly those related to Desulfatirhabdium butyrativorans. Methanogens were dominant archaea and possibly were present at higher relative abundances than SRM in some BCRs. Both hydrogenotrophic and acetoclastic types were present. There were no strong negative or positive co-occurrence correlations of methanogen and SRM taxa. Knowing which SRM inhabit successfully operating BCRs allows practitioners to target these phylogenetic groups when selecting inoculum for future operations. -

Community Structure Analysis �9
Biodegradation of Polystyrene Foam by the Microorganisms from Landfill Pat Pataranutaporn ! Assistant prof. Savaporn Supaphol prof. Amornrat Phongdara Sureeporn Nualkaew Hi, I would like to invite you to take a look on my research Pat Introduction !3 “Styrofoam” Polystyrene Disadvantage Physical Properties ! • chemical formula is (C8H8)n • Non-biodegradable in the environment • monomer styrene • Made from non-renewable petroleum products • Thermoplastic • Chronic, low-level exposure risks undetermined • blowing agents Introduction !4 Bacteria nutritional requirements ! ‣ Energy source Biodegradation ‣ Carbon source Possibly work? ‣ Nitrogen source ‣ Minerals ‣ Water ‣ Growth factors Polystyrene structure http://faculty.ccbcmd.edu/courses/bio141/ lecguide/unit6/metabolism/growth/factors.html Introduction !5 Aims of the research ‣To identify the microbe that able to growth in the condition that polystyrene is a sole carbon source ! ‣To study the changing of microbe community structure in the selective culture which polystyrene is a sole carbon source ! ‣To observe the biodegradability of polystyrene To analyse the by product of polystyrene after degradation Methodology Methodology !7 Agar cultivation Community fingerprint 2 16s Ribosomal RNA Microbe Screening months later identification sampling Cultivation Molecular cloning Phylogenetic tree Degradability observation (SEM) Methodology Microbe sampling & cultivation !8 Agar cultivation Community fingerprint 2 16s Ribosomal RNA Microbe Screening months later identification sampling Cultivation -

1 Microbial Transformations of Organic Chemicals in Produced Fluid From
Microbial transformations of organic chemicals in produced fluid from hydraulically fractured natural-gas wells Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Morgan V. Evans Graduate Program in Environmental Science The Ohio State University 2019 Dissertation Committee Professor Paula Mouser, Advisor Professor Gil Bohrer, Co-Advisor Professor Matthew Sullivan, Member Professor Ilham El-Monier, Member Professor Natalie Hull, Member 1 Copyrighted by Morgan Volker Evans 2019 2 Abstract Hydraulic fracturing and horizontal drilling technologies have greatly improved the production of oil and natural-gas from previously inaccessible non-permeable rock formations. Fluids comprised of water, chemicals, and proppant (e.g., sand) are injected at high pressures during hydraulic fracturing, and these fluids mix with formation porewaters and return to the surface with the hydrocarbon resource. Despite the addition of biocides during operations and the brine-level salinities of the formation porewaters, microorganisms have been identified in input, flowback (days to weeks after hydraulic fracturing occurs), and produced fluids (months to years after hydraulic fracturing occurs). Microorganisms in the hydraulically fractured system may have deleterious effects on well infrastructure and hydrocarbon recovery efficiency. The reduction of oxidized sulfur compounds (e.g., sulfate, thiosulfate) to sulfide has been associated with both well corrosion and souring of natural-gas, and proliferation of microorganisms during operations may lead to biomass clogging of the newly created fractures in the shale formation culminating in reduced hydrocarbon recovery. Consequently, it is important to elucidate microbial metabolisms in the hydraulically fractured ecosystem. -

Which Organisms Are Used for Anti-Biofouling Studies
Table S1. Semi-systematic review raw data answering: Which organisms are used for anti-biofouling studies? Antifoulant Method Organism(s) Model Bacteria Type of Biofilm Source (Y if mentioned) Detection Method composite membranes E. coli ATCC25922 Y LIVE/DEAD baclight [1] stain S. aureus ATCC255923 composite membranes E. coli ATCC25922 Y colony counting [2] S. aureus RSKK 1009 graphene oxide Saccharomycetes colony counting [3] methyl p-hydroxybenzoate L. monocytogenes [4] potassium sorbate P. putida Y. enterocolitica A. hydrophila composite membranes E. coli Y FESEM [5] (unspecified/unique sample type) S. aureus (unspecified/unique sample type) K. pneumonia ATCC13883 P. aeruginosa BAA-1744 composite membranes E. coli Y SEM [6] (unspecified/unique sample type) S. aureus (unspecified/unique sample type) graphene oxide E. coli ATCC25922 Y colony counting [7] S. aureus ATCC9144 P. aeruginosa ATCCPAO1 composite membranes E. coli Y measuring flux [8] (unspecified/unique sample type) graphene oxide E. coli Y colony counting [9] (unspecified/unique SEM sample type) LIVE/DEAD baclight S. aureus stain (unspecified/unique sample type) modified membrane P. aeruginosa P60 Y DAPI [10] Bacillus sp. G-84 LIVE/DEAD baclight stain bacteriophages E. coli (K12) Y measuring flux [11] ATCC11303-B4 quorum quenching P. aeruginosa KCTC LIVE/DEAD baclight [12] 2513 stain modified membrane E. coli colony counting [13] (unspecified/unique colony counting sample type) measuring flux S. aureus (unspecified/unique sample type) modified membrane E. coli BW26437 Y measuring flux [14] graphene oxide Klebsiella colony counting [15] (unspecified/unique sample type) P. aeruginosa (unspecified/unique sample type) graphene oxide P. aeruginosa measuring flux [16] (unspecified/unique sample type) composite membranes E. -

Supplementary File 1
Supplementary File 1. Unlocking survival mechanisms for metal and oxidative stress in the extremely acidophilic, halotolerant Acidihalobacter genus Himel Nahreen Khaleque1,5, Homayoun Fathollazadeh1, Carolina González2,4, Raihan Shafique1, Anna H. Kaksonen5, David S. Holmes2,3,4 and Elizabeth L.J. Watkin1* 1School of Pharmacy and Biomedical Sciences, Curtin University, Perth, Australia; 2Center for Bioinformatics and Genome Biology, Fundacion Ciencia y Vida, Santiago, Chile; 3Universidad San Sebastian, Santiago Chile; 4Centro de Genómica y Bioinformática, Facultad de Ciencias, Universidad Mayor, Santiago, Chile; and 5CSIRO Land and Water, Floreat, Australia *Correspondence: [email protected]; Tel.: (+61 8 92662955) Table S1. BLASTx analysis of the Acidihalobacter yilgarnensis F5T operon containing mobile genetic elements and copper resistance genes. Next best hits in red = genes with similarity to orthologs in acidophiles; green = genes with similarity to orthologs in neutrophilic halophilic and halotolerant organisms or to other non-acidophiles, black = unique to Acidihalobacter, blue = genes with similarity to orthologs found across the domain Bacteria. peg. RAST annotation NCBI annotation Ident (%) to NCBI accession Next best hit (NBH) Ident NCBI accession No Acidhalocbacter (%) to sp. NBH 2773 FIG00761799: DUF4396 domain- 100% WP_070080076.1 DUF4396 domain- 55.02% WP_110138179.1 membrane protein containing protein containing protein CDS [Acidihalobacter [Acidiferrobacter sp. prosperus] SPIII_3] 2774 Hypothetical protein hypothetical -

Textile Waste and Microplastic Induce Activity and Development of Unique
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.08.939876; this version posted February 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Textile waste and microplastic induce activity and 2 development of unique hydrocarbon-degrading marine 3 bacterial communities 4 5 Elsa B. Girard1, Melanie Kaliwoda2, Wolfgang W. Schmahl1,2,3, Gert Wörheide1,3,4 and 6 William D. Orsi1,3* 7 8 1 Department of Earth and Environmental Sciences, Ludwig-Maximilians-Universität München, 9 80333 Munich, Germany 10 2 SNSB - Mineralogische Staatssammlung München, 80333 München, Germany 11 3 GeoBio-CenterLMU, Ludwig-Maximilians-Universität München, 80333 Munich, Germany 12 4 SNSB - Bayerische Staatssammlung für Paläontologie und Geologie, 80333 Munich, Germany 13 *Corresponding author (e-mail: [email protected]) 14 15 16 17 18 KEYWORDS 19 Microplastic, Fiber, Hydrocarbon-degrading bacteria, Microbial community, Pollution 20 21 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.08.939876; this version posted February 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 22 ABSTRACT 23 Biofilm-forming microbial communities on plastics and textile fibers are of growing interest since 24 they have potential to contribute to disease outbreaks and material biodegradability in the 25 environment. -

A Web Tool for Hydrogenase Classification and Analysis
bioRxiv preprint doi: https://doi.org/10.1101/061994; this version posted September 16, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 HydDB: A web tool for hydrogenase 2 classification and analysis 3 Dan Søndergaarda, Christian N. S. Pedersena, Chris Greeningb, c* 4 5 a Aarhus University, Bioinformatics Research Centre, C.F. Møllers Allé 8, Aarhus 6 DK-8000, Denmark 7 b The Commonwealth Scientific and Industrial Research Organisation, Land and 8 Water Flagship, Clunies Ross Street, Acton, ACT 2060, Australia 9 c Monash University, School of Biological Sciences, Clayton, VIC 2800, Australia 10 11 Correspondence: 12 Dr Chris Greening ([email protected]), Monash University, School of 13 Biological Sciences, Clayton, VIC 2800, Australia 14 Dan Søndergaard ([email protected]), Aarhus University, Bioinformatics Research 15 Centre, C.F. Møllers Allé 8, Aarhus DK-8000, Denmark 1 bioRxiv preprint doi: https://doi.org/10.1101/061994; this version posted September 16, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 16 Abstract 17 H2 metabolism is proposed to be the most ancient and diverse mechanism of 18 energy-conservation. The metalloenzymes mediating this metabolism, 19 hydrogenases, are encoded by over 60 microbial phyla and are present in all major 20 ecosystems. -

A Noval Investigation of Microbiome from Vermicomposting Liquid Produced by Thai Earthworm, Perionyx Sp
International Journal of Agricultural Technology 2021Vol. 17(4):1363-1372 Available online http://www.ijat-aatsea.com ISSN 2630-0192 (Online) A novel investigation of microbiome from vermicomposting liquid produced by Thai earthworm, Perionyx sp. 1 Kraisittipanit, R.1,2, Tancho, A.2,3, Aumtong, S.3 and Charerntantanakul, W.1* 1Program of Biotechnology, Faculty of Science, Maejo University, Chiang Mai, Thailand; 2Natural Farming Research and Development Center, Maejo University, Chiang Mai, Thailand; 3Faculty of Agricultural Production, Maejo University, Thailand. Kraisittipanit, R., Tancho, A., Aumtong, S. and Charerntantanakul, W. (2021). A noval investigation of microbiome from vermicomposting liquid produced by Thai earthworm, Perionyx sp. 1. International Journal of Agricultural Technology 17(4):1363-1372. Abstract The whole microbiota structure in vermicomposting liquid derived from Thai earthworm, Perionyx sp. 1 was estimated. It showed high richness microbial species and belongs to 127 species, separated in 3 fungal phyla (Ascomycota, Basidiomycota, Mucoromycota), 1 Actinomycetes and 16 bacterial phyla (Acidobacteria, Armatimonadetes, Bacteroidetes, Balneolaeota, Candidatus, Chloroflexi, Deinococcus, Fibrobacteres, Firmicutes, Gemmatimonadates, Ignavibacteriae, Nitrospirae, Planctomycetes, Proteobacteria, Tenericutes and Verrucomicrobia). The OTUs data analysis revealed the highest taxonomic abundant ratio in bacteria and fungi belong to Proteobacteria (70.20 %) and Ascomycota (5.96 %). The result confirmed that Perionyx sp. 1