The Discovery of Hepatitis C Virus
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Immunogenicity and Efficacy Testing in Chimpanzees of an Oral Hepatitis B Vaccine Based on Live Recombinant Adenovirus
Proc. Natl. Acad. Sci. USA Vol. 86, pp. 6763-6767, September 1989 Medical Sciences Immunogenicity and efficacy testing in chimpanzees of an oral hepatitis B vaccine based on live recombinant adenovirus (adenovirus animal model/adenovirus-vectored vacdnes) MICHAEL D. LUBECK*, ALAN R. DAVIS*, MURTY CHENGALVALA*, ROBERT J. NATUK*, JOHN E. MORIN*, KATHERINE MOLNAR-KIMBER*, BRUCE B. MASON*, BHEEM M. BHAT*, SATOSHI MIZUTANI*, PAUL P. HUNG*, AND ROBERT H. PURCELLO *Wyeth-Ayerst Research, Biotechnology and Microbiology Division, P.O. Pox 8299, Philadelphia, PA 19101; and tLaboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892 Contributed by Robert H. Purcell, May 30, 1989 ABSTRACT As a major cause of acute and chronic liver which, because oftheir wide prior usage, are good candidates disease as well as hepatocellular carcinoma, hepatitis B virus as vectors. Other less well-characterized small animal models (HBV) continues to pose significant health problems world- for human adenoviruses have been occasionally reported wide. Recombinant hepatitis B vaccines based on adenovirus (11-13) but are likewise nonpermissive for Ad4 and Ad7 vectors have been developed to address global needs for infections (unpublished data). An early study of animal effective control of hepatitits B infection. Although consider- species including nonhuman primates indicated that human able progress has been made in the construction ofrecombinant adenoviruses do not induce acute respiratory disease in adenoviruses that express large amounts of HBV gene prod- monkeys or chimpanzees following intranasal inoculations ucts, preclinical immunogenicity and efficacy testing of candi- (14). Serological data, however, indicated that such experi- date vaccines has remained difficult due to the lack ofa suitable mental infections occasionally induced anti-adenovirus anti- animal model. -

Hepatitis B Fast Facts Everything You Need to Know in 2 Minutes Or Less!
Hepatitis B Foundation Cause for a Cure www.hepb.org Hepatitis B Fast Facts Everything you need to know in 2 minutes or less! Hepatitis B is the most common serious liver infection in the world. It is caused by the hepatitis B virus (HBV) that attacks liver cells and can lead to liver failure, cirrhosis (scarring) or cancer of the liver. The virus is transmitted through contact with blood and bodily fluids that contain blood. Most people are able to fight off the hepatitis B infection and clear the virus from their blood. This may take up to six months. While the virus is present in their blood, infected people can pass the virus on to others. Approximately 5-10% of adults, 30-50% of children, and 90% of babies will not get rid of the virus and will develop chronic infection. Chronically infected people can pass the virus on to others and are at increased risk for liver problems later in life. The hepatitis B virus is 100 times more infectious than the AIDS virus. Yet, hepatitis B can be pre- vented with a safe and effective vaccine. For the 400 million people worldwide who are chronically infected with hepatitis B, the vaccine is of no use. However, there are promising new treatments for those who live with chronic hepatitis B. In the World: • This year alone, 10 to 30 million people will become infected with the hepatitis B virus (HBV). • The World Health Organization estimates that 400 million people worldwide are already chronically infected with hepatitis B. -

Prevention & Control of Viral Hepatitis Infection
Prevention & Control of Viral Hepatitis Infection: A Strategy for Global Action © World Health Organization 2011. All rights reserved. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by WHO to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either express or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. Table of contents Disease burden 02 What is viral hepatitis? 05 Prevention & control: a tailored approach 06 Global Achievements 08 Remaining challenges 10 World Health Assembly: a mandate for comprehensive prevention & control 13 WHO goals and strategy -

The Role of Hepatitis C Virus in Hepatocellular Carcinoma U
Viruses in cancer cell plasticity: the role of hepatitis C virus in hepatocellular carcinoma U. Hibner, D. Gregoire To cite this version: U. Hibner, D. Gregoire. Viruses in cancer cell plasticity: the role of hepatitis C virus in hepato- cellular carcinoma. Contemporary Oncology, Termedia Publishing House, 2015, 19 (1A), pp.A62–7. 10.5114/wo.2014.47132. hal-02187396 HAL Id: hal-02187396 https://hal.archives-ouvertes.fr/hal-02187396 Submitted on 2 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License Review Viruses are considered as causative agents of a significant proportion of human cancers. While the very Viruses in cancer cell plasticity: stringent criteria used for their clas- sification probably lead to an under- estimation, only six human viruses the role of hepatitis C virus are currently classified as oncogenic. In this review we give a brief histor- in hepatocellular carcinoma ical account of the discovery of on- cogenic viruses and then analyse the mechanisms underlying the infectious causes of cancer. We discuss viral strategies that evolved to ensure vi- Urszula Hibner1,2,3, Damien Grégoire1,2,3 rus propagation and spread can alter cellular homeostasis in a way that increases the probability of oncogen- 1Institut de Génétique Moléculaire de Montpellier, CNRS, UMR 5535, Montpellier, France ic transformation and acquisition of 2Université Montpellier 2, Montpellier, France stem cell phenotype. -

Entry of Hepatitis B and C Viruses
VIRAL HEPATITIS FORUM Getting Close Viralto Eradication Hepatitis Forum I. Basic Getting Research Close to Eradication I. Basic Research Entry of Hepatitis B and C Viruses Seungtaek Kim Severance Biomedical Science Institute, Institute of Gastroenterology, Department of Internal Medicine, Yonsei University Col- lege of Medicine, Seoul, Korea B형과 C형 간염 바이러스에 대한 최근의 분자, 세포생물학적인 발전은 간세포를 특이적으로 감염시키는 이들 바이러스에 대한 세포 수용체의 발굴과 더불어 그들의 작용 기전에 대해 더 자세한 정보들을 제공해주고 있다. 특히 C형 간염 바이러스의 경우, 간세포의 서로 다른 곳에 위치한 세포 수용체들이 바이러스의 세포 진입시에 바이러스 표면의 당단백질과 어떤 방식으로 서로 상호 작용하며 세포 내 신호 전달 과정을 거쳐 세포 안으로 들어오게 되는지 그 기전들이 서서히 드러나고 있다. 한편, B형 간염 바이러스의 경우, 오랫동안 밝혀내지 못했던 이 바이러스의 세포 수용체인 NTCP를 최근 발굴하게 됨으로써 세포 진입에 관한 연구에 획기적인 계기를 마련하게 되었으며 동시에 이를 저해할 수 있는 새로운 항바이러스제의 개발도 활기를 띠게 되었다. 임상적으로 매우 중요한 이 두 바이러스의 세포 진입에 관한 연구는 앞으로도 매우 활발하게 이루어질 것으로 기대된다. Keywords: B형 간염 바이러스, C형 간염 바이러스, 세포 진입, 신호 전달, NTCP There are five hepatitis viruses although their classes, genomes, and modes of transmission are different from each other. Of these, hepatitis B virus (HBV) and hepatitis C virus (HCV) are the most dangerous, life-threatening pathogens, which are also responsible for 80-90% of hepatocellular carcinoma. HBV belongs to hepadnaviridae (family) and it has double-strand DNA as its genome, however, its replication occurs via reverse transcription like retrovirus replication. In contrast, HCV belongs to flaviviridae (family) and has positive-sense, single-strand RNA as its genome. -

Symposium on Viral Membrane Proteins
Viral Membrane Proteins ‐ Shanghai 2011 交叉学科论坛 Symposium for Advanced Studies 第二十七期:病毒离子通道蛋白的结构与功能研讨会 Symposium on Viral Membrane Proteins 主办单位:中国科学院上海交叉学科研究中心 承办单位:上海巴斯德研究所 1 Viral Membrane Proteins ‐ Shanghai 2011 Symposium on Viral Membrane Proteins Shanghai Institute for Advanced Studies, CAS Institut Pasteur of Shanghai,CAS 30.11. – 2.12 2011 Shanghai, China 2 Viral Membrane Proteins ‐ Shanghai 2011 Schedule: Wednesday, 30th of November 2011 Morning Arrival Thursday, 1st of December 2011 8:00 Arrival 9:00 Welcome Bing Sun, Co-Director, Pasteur Institute Shanghai 9: 10 – 9:35 Bing Sun, Pasteur Institute Shanghai Ion channel study and drug target fuction research of coronavirus 3a like protein. 9:35 – 10:00 Tim Cross, Tallahassee, USA The proton conducting mechanism and structure of M2 proton channel in lipid bilayers. 10:00 – 10:25 Shy Arkin, Jerusalem, IL A backbone structure of SARS Coronavirus E protein based on Isotope edited FTIR, X-ray reflectivity and biochemical analysis. 10:20 – 10:45 Coffee Break 10:45 – 11:10 Rainer Fink, Heidelberg, DE Elektromechanical coupling in muscle: a viral target? 11:10 – 11:35 Yechiel Shai, Rehovot, IL The interplay between HIV1 fusion peptide, the transmembrane domain and the T-cell receptor in immunosuppression. 11:35 – 12:00 Christoph Cremer, Mainz and Heidelberg University, DE Super-resolution Fluorescence imaging of cellular and viral nanostructures. 12:00 – 13:30 Lunch Break 3 Viral Membrane Proteins ‐ Shanghai 2011 13:30 – 13:55 Jung-Hsin Lin, National Taiwan University Robust Scoring Functions for Protein-Ligand Interactions with Quantum Chemical Charge Models. 13:55 – 14:20 Martin Ulmschneider, Irvine, USA Towards in-silico assembly of viral channels: the trials and tribulations of Influenza M2 tetramerization. -

Opportunistic Intruders: How Viruses Orchestrate ER Functions to Infect Cells
REVIEWS Opportunistic intruders: how viruses orchestrate ER functions to infect cells Madhu Sudhan Ravindran*, Parikshit Bagchi*, Corey Nathaniel Cunningham and Billy Tsai Abstract | Viruses subvert the functions of their host cells to replicate and form new viral progeny. The endoplasmic reticulum (ER) has been identified as a central organelle that governs the intracellular interplay between viruses and hosts. In this Review, we analyse how viruses from vastly different families converge on this unique intracellular organelle during infection, co‑opting some of the endogenous functions of the ER to promote distinct steps of the viral life cycle from entry and replication to assembly and egress. The ER can act as the common denominator during infection for diverse virus families, thereby providing a shared principle that underlies the apparent complexity of relationships between viruses and host cells. As a plethora of information illuminating the molecular and cellular basis of virus–ER interactions has become available, these insights may lead to the development of crucial therapeutic agents. Morphogenesis Viruses have evolved sophisticated strategies to establish The ER is a membranous system consisting of the The process by which a virus infection. Some viruses bind to cellular receptors and outer nuclear envelope that is contiguous with an intri‑ particle changes its shape and initiate entry, whereas others hijack cellular factors that cate network of tubules and sheets1, which are shaped by structure. disassemble the virus particle to facilitate entry. After resident factors in the ER2–4. The morphology of the ER SEC61 translocation delivering the viral genetic material into the host cell and is highly dynamic and experiences constant structural channel the translation of the viral genes, the resulting proteins rearrangements, enabling the ER to carry out a myriad An endoplasmic reticulum either become part of a new virus particle (or particles) of functions5. -

Rational Engineering of HCV Vaccines for Humoral Immunity
viruses Review To Include or Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity Felicia Schlotthauer 1,2,†, Joey McGregor 1,2,† and Heidi E Drummer 1,2,3,* 1 Viral Entry and Vaccines Group, Burnet Institute, Melbourne, VIC 3004, Australia; [email protected] (F.S.); [email protected] (J.M.) 2 Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC 3000, Australia 3 Department of Microbiology, Monash University, Clayton, VIC 3800, Australia * Correspondence: [email protected]; Tel.: +61-392-822-179 † These authors contributed equally to this work. Abstract: Direct-acting antiviral agents have proven highly effective at treating existing hepatitis C infections but despite their availability most countries will not reach the World Health Organization targets for elimination of HCV by 2030. A prophylactic vaccine remains a high priority. Whilst early vaccines focused largely on generating T cell immunity, attention is now aimed at vaccines that gen- erate humoral immunity, either alone or in combination with T cell-based vaccines. High-resolution structures of hepatitis C viral glycoproteins and their interaction with monoclonal antibodies isolated from both cleared and chronically infected people, together with advances in vaccine technologies, provide new avenues for vaccine development. Keywords: glycoprotein E2; vaccine development; humoral immunity Citation: Schlotthauer, F.; McGregor, J.; Drummer, H.E To Include or 1. Introduction Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity. The development of direct-acting antiviral agents (DAA) with their ability to cure Viruses 2021, 13, 805. https:// infection in >95% of those treated was heralded as the key to eliminating hepatitis C globally. -

Hepatitis B? HEPATITIS B Hepatitis B Is a Contagious Liver Disease That Results from Infection with the Hepatitis B Virus
What is Hepatitis B? HEPATITIS B Hepatitis B is a contagious liver disease that results from infection with the Hepatitis B virus. When first infected, a person can develop Are you at risk? an “acute” infection, which can range in severity from a very mild illness with few or no symptoms to a serious condition requiring hospitalization. Acute Hepatitis B refers to the first 6 months after someone is exposed to the Hepatitis B virus. Some people are able to fight the infection and clear the virus. For others, the infection remains and leads to a “chronic,” or lifelong, illness. Chronic Hepatitis B refers to the illness that occurs when the Hepatitis B virus remains in a person’s body. Over time, the infection can cause serious health problems. How is Hepatitis B spread? Hepatitis B is usually spread when blood, semen, or other body fluids from a person infected with the Hepatitis B virus enter the body of someone who is not infected. This can happen through having sex with an infected partner; sharing needles, syringes, or other injection drug equipment; or from direct contact with the blood or open sores of an infected person. Hepatitis B can also be passed from an infected mother to her baby at birth. Who should be tested for Hepatitis B? Approximately 1.2 million people in the United States and 350 million people worldwide have Hepatitis B. Testing for Hepatitis B is recommended for certain groups of people, including: Most are unaware of their infection. ■ People born in Asia, Africa, and other regions with moderate or high rates Is Hepatitis B common? of Hepatitis B (see map) Yes. -
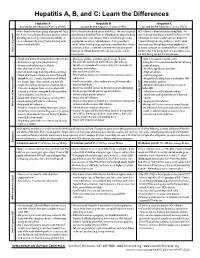
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

(12) United States Patent (10) Patent No.: US 7,790,366 B1 Houghton Et Al
US007790366B1 (12) United States Patent (10) Patent No.: US 7,790,366 B1 Houghton et al. (45) Date of Patent: *Sep. 7, 2010 (54) NANBV DIAGNOSTICS AND VACCINES 4,683, 195 A 7, 1987 Mullis et al. ................... 435/6 4,683.202 A 7, 1987 Mullis et al. ............... 435,912 (75) Inventors: Michael Houghton, Danville, CA (US); 4,702,909. A 10/1987 Villarejos et al. .............. 435/5 GeorgeQui-Lim Kuo, Choo, San El Francisco, Cerrito, CA CA (US); (US) 4,707.439 A * 1 1/1987 Seto .............................. 435/5 4,722,897 A * 2/1988 Soeda et al. ............... 435/69.1 (73) Assignee: Novartis Vaccines and Diagnostics, 4,769,326 A * 9/1988 Rutter ....................... 435.68.1 Inc., Emeryville, CA (US) 4,870,026 A * 9/1989 Wands ........................ 436,538 (*) Notice: Subject to any disclaimer, the term of this 4,891.313 A 1/1990 Berger et al. ............... 435/7.94 patent is extended or adjusted under 35 5,077, 193 A 12, 1991 Mishiro et al. ................. 435/5 U.S.C. 154(b) by 0 days. 5,101,024 A * 3/1992 Kudo et al. .............. 536,23.53 5,106,726 A 4/1992 Wang ............................ 435/5 This patent is Subject to a terminal dis- 5, 191,064 A 3/1993 Arima et al. ................ 530/324 claimer. 5,218,099 A 6/1993 Reyes et al. .............. 536,23.72 5,268,278 A * 12/1993 Canosi et al. .............. 435/69.7 (21) Appl. No.: 08/441,443 5,310,893 A * 5/1994 Erlich et al. ............. 536, 24.31 (22) Filed: May 15, 1995 5,372,928 A 12/1994 Miyamura 5,436,126 A 7/1995 Wang ........................... -

Understanding Human Astrovirus from Pathogenesis to Treatment
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 6-2020 Understanding Human Astrovirus from Pathogenesis to Treatment Virginia Hargest University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Diseases Commons, Medical Sciences Commons, and the Viruses Commons Recommended Citation Hargest, Virginia (0000-0003-3883-1232), "Understanding Human Astrovirus from Pathogenesis to Treatment" (2020). Theses and Dissertations (ETD). Paper 523. http://dx.doi.org/10.21007/ etd.cghs.2020.0507. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Understanding Human Astrovirus from Pathogenesis to Treatment Abstract While human astroviruses (HAstV) were discovered nearly 45 years ago, these small positive-sense RNA viruses remain critically understudied. These studies provide fundamental new research on astrovirus pathogenesis and disruption of the gut epithelium by induction of epithelial-mesenchymal transition (EMT) following astrovirus infection. Here we characterize HAstV-induced EMT as an upregulation of SNAI1 and VIM with a down regulation of CDH1 and OCLN, loss of cell-cell junctions most notably at 18 hours post-infection (hpi), and loss of cellular polarity by 24 hpi. While active transforming growth factor- (TGF-) increases during HAstV infection, inhibition of TGF- signaling does not hinder EMT induction. However, HAstV-induced EMT does require active viral replication.