Establishment of Anthoceros Agrestis As a Model Species for Studying the Biology of Hornworts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novelties in the Hornwort Flora of Croatia and Southeast Europe
cryptogamie Bryologie 2019 ● 40 ● 22 DIRECTEUR DE LA PUBLICATION : Bruno David, Président du Muséum national d’Histoire naturelle RÉDACTEURS EN CHEF / EDITORS-IN-CHIEF : Denis LAMY ASSISTANTS DE RÉDACTION / ASSISTANT EDITORS : Marianne SALAÜN ([email protected]) MISE EN PAGE / PAGE LAYOUT : Marianne SALAÜN RÉDACTEURS ASSOCIÉS / ASSOCIATE EDITORS Biologie moléculaire et phylogénie / Molecular biology and phylogeny Bernard GOFFINET Department of Ecology and Evolutionary Biology, University of Connecticut (United States) Mousses d’Europe / European mosses Isabel DRAPER Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) Francisco LARA GARCÍA Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) Mousses d’Afrique et d’Antarctique / African and Antarctic mosses Rysiek OCHYRA Laboratory of Bryology, Institute of Botany, Polish Academy of Sciences, Krakow (Pologne) Bryophytes d’Asie / Asian bryophytes Rui-Liang ZHU School of Life Science, East China Normal University, Shanghai (China) Bioindication / Biomonitoring Franck-Olivier DENAYER Faculté des Sciences Pharmaceutiques et Biologiques de Lille, Laboratoire de Botanique et de Cryptogamie, Lille (France) Écologie des bryophytes / Ecology of bryophyte Nagore GARCÍA MEDINA Department of Biology (Botany), and Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) COUVERTURE / COVER : Extraits d’éléments de la Figure 2 / Extracts of -

Phytotaxa, a Synthesis of Hornwort Diversity
Phytotaxa 9: 150–166 (2010) ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ Article PHYTOTAXA Copyright © 2010 • Magnolia Press ISSN 1179-3163 (online edition) A synthesis of hornwort diversity: Patterns, causes and future work JUAN CARLOS VILLARREAL1 , D. CHRISTINE CARGILL2 , ANDERS HAGBORG3 , LARS SÖDERSTRÖM4 & KAREN SUE RENZAGLIA5 1Department of Ecology and Evolutionary Biology, University of Connecticut, 75 North Eagleville Road, Storrs, CT 06269; [email protected] 2Centre for Plant Biodiversity Research, Australian National Herbarium, Australian National Botanic Gardens, GPO Box 1777, Canberra. ACT 2601, Australia; [email protected] 3Department of Botany, The Field Museum, 1400 South Lake Shore Drive, Chicago, IL 60605-2496; [email protected] 4Department of Biology, Norwegian University of Science and Technology, N-7491 Trondheim, Norway; [email protected] 5Department of Plant Biology, Southern Illinois University, Carbondale, IL 62901; [email protected] Abstract Hornworts are the least species-rich bryophyte group, with around 200–250 species worldwide. Despite their low species numbers, hornworts represent a key group for understanding the evolution of plant form because the best–sampled current phylogenies place them as sister to the tracheophytes. Despite their low taxonomic diversity, the group has not been monographed worldwide. There are few well-documented hornwort floras for temperate or tropical areas. Moreover, no species level phylogenies or population studies are available for hornworts. Here we aim at filling some important gaps in hornwort biology and biodiversity. We provide estimates of hornwort species richness worldwide, identifying centers of diversity. We also present two examples of the impact of recent work in elucidating the composition and circumscription of the genera Megaceros and Nothoceros. -

Anthocerotophyta
Glime, J. M. 2017. Anthocerotophyta. Chapt. 2-8. In: Glime, J. M. Bryophyte Ecology. Volume 1. Physiological Ecology. Ebook 2-8-1 sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 5 June 2020 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 2-8 ANTHOCEROTOPHYTA TABLE OF CONTENTS Anthocerotophyta ......................................................................................................................................... 2-8-2 Summary .................................................................................................................................................... 2-8-10 Acknowledgments ...................................................................................................................................... 2-8-10 Literature Cited .......................................................................................................................................... 2-8-10 2-8-2 Chapter 2-8: Anthocerotophyta CHAPTER 2-8 ANTHOCEROTOPHYTA Figure 1. Notothylas orbicularis thallus with involucres. Photo by Michael Lüth, with permission. Anthocerotophyta These plants, once placed among the bryophytes in the families. The second class is Leiosporocerotopsida, a Anthocerotae, now generally placed in the phylum class with one order, one family, and one genus. The genus Anthocerotophyta (hornworts, Figure 1), seem more Leiosporoceros differs from members of the class distantly related, and genetic evidence may even present -

Morphology, Anatomy and Reproduction of Anthoceros
Morphology, Anatomy and Reproduction of Anthoceros - INDRESH KUMAR PANDEY Taxonomic Position of Anthoceros Class- Anthocerotopsida Single order Anthocerotales Two families Anthocerotaceae Notothylaceae Representative Genus Anthoceros Notothylus General features of Anthocerotopsida • Forms an isolated evolutionary line • Sometimes considered independent from Bryophytes and placed in division Anthocerophyta • Called as Hornworts due to horn like structure of sporophyte • Commonly recognised genera includes Anthoceros, Megaceros, Nothothylus, Dendroceros Anthoceros :Habitat & Distribution • Cosmopolitan • Mainly in temperate & tropical regions • More than 200 species, 25 sp. Recorded from India. • Mostly grows in moist shady places, sides of ditches or in moist hollows among rocks • Few species grow on decaying wood. • Three common Indian species- A. erectus, A. crispulus, A. himalayensis Anthoceros: Morphology Dorsal surface Ventral surface Rhizoids (smooth walled) Thallus showing tubers External features • Thallus (gametophyte)- small, dark green, dorsiventral, prostrate, branched or lobed • No midrib, spongy due to presence of underlying mucilaginous ducts • Dorsal surface varies from species to species Smooth- A. laevis Velvety- A. crispulus Rough- A. fusiformis • Smooth walled rhizoid on ventral surface • Rounded bluish green thickened area on ventral surface- Nostoc colonies Internal structure Vertical Transverse Section- Diagrammatic Vertical Transverse Section- Cellular Internal Structure • Simple, without cellular differentiation -

About the Book the Format Acknowledgments
About the Book For more than ten years I have been working on a book on bryophyte ecology and was joined by Heinjo During, who has been very helpful in critiquing multiple versions of the chapters. But as the book progressed, the field of bryophyte ecology progressed faster. No chapter ever seemed to stay finished, hence the decision to publish online. Furthermore, rather than being a textbook, it is evolving into an encyclopedia that would be at least three volumes. Having reached the age when I could retire whenever I wanted to, I no longer needed be so concerned with the publish or perish paradigm. In keeping with the sharing nature of bryologists, and the need to educate the non-bryologists about the nature and role of bryophytes in the ecosystem, it seemed my personal goals could best be accomplished by publishing online. This has several advantages for me. I can choose the format I want, I can include lots of color images, and I can post chapters or parts of chapters as I complete them and update later if I find it important. Throughout the book I have posed questions. I have even attempt to offer hypotheses for many of these. It is my hope that these questions and hypotheses will inspire students of all ages to attempt to answer these. Some are simple and could even be done by elementary school children. Others are suitable for undergraduate projects. And some will take lifelong work or a large team of researchers around the world. Have fun with them! The Format The decision to publish Bryophyte Ecology as an ebook occurred after I had a publisher, and I am sure I have not thought of all the complexities of publishing as I complete things, rather than in the order of the planned organization. -
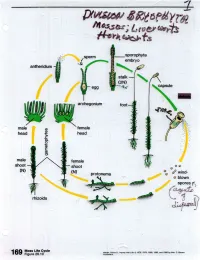
02-Bryophyta-2.Pdf
498 INTRODUCTORY PLANT SCIENCE layer. In certain species, the capsule con til1Ues to grow as long as the gametophyte lives. The presence of a meristcm in A11tlioceros and an aerating system complete with sto mata may indicate that Antlwceros evolved from ancestors with even ·larger· and more 1. Civ complex sporophytes. These features may Bryophyt be vestiges from a more complex ancestral. 2. Wh sporophyte. that ti alga ? 3. De� ORIGIN AND RELATIONSHIPS OF THE Bryophyt, BRYOPHYTA 4._ characteri Little is known with certainty about the r are small origin and evolution of the bryophytes. The ' tall. ( B) fossil record is too fragmenta to enable • ry s true roots us to trace their evolutionary history. Frag ,, sperms ar mentary remnants of thallose liverworts, spore idium and mother\4'/ttN) which resemble present-day liverworts, have cell Jar arch� been foum! in rocks of Carboniferous age, bryophyt as have structures that may be remains of more com mosses. all bryoph The immediate ancestors of the bryo bryo whicl phytes were probably more complex plants from the have an i than present-day forms. In other words, the gametophy evolutionary tendency has been one of re- with a dip1 duction instead of increased complexity. •• • • • spores If evolution has progressed . from more • . •, .... • s. __ complex sporophytes to those of simpler • . vascular p Anthoceros . form, the sporophyte of wou1d • • green algat be considered more ancient than that of • (D) red Marchantia or Riccia. Because Riccia has Fig. 35-16. longitudinal section of the spo 6. How the most reduced sporophyte, it would be t·:i;,hyte of Anthoceros. -

Chapter 3-1 Sexuality: Sexual Strategies Janice M
Glime, J. M. and Bisang, I. 2017. Sexuality: Sexual Strategies. Chapt. 3-1. In: Glime, J. M. Bryophyte Ecology. Volume 1. 3-1-1 Physiological Ecology. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 2 April 2017 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 3-1 SEXUALITY: SEXUAL STRATEGIES JANICE M. GLIME AND IRENE BISANG TABLE OF CONTENTS Expression of Sex............................................................................................................................................... 3-1-2 Unisexual and Bisexual Taxa............................................................................................................................. 3-1-2 Sex Chromosomes....................................................................................................................................... 3-1-6 An unusual Y Chromosome........................................................................................................................ 3-1-7 Gametangial Arrangement.......................................................................................................................... 3-1-8 Origin of Bisexuality in Bryophytes ................................................................................................................ 3-1-11 Monoicy as a Derived/Advanced Character.............................................................................................. 3-1-11 Anthocerotophyta and Multiple Reversals............................................................................................... -

Anthocerotophyta) of Colombia
BOTANY https://dx.doi.org/10.15446/caldasia.v40n2.71750 http://www.revistas.unal.edu.co/index.php/cal Caldasia 40(2):262-270. Julio-diciembre 2018 Key to hornworts (Anthocerotophyta) of Colombia Clave para Antocerotes (Anthocerotophyta) de Colombia S. ROBBERT GRADSTEIN Muséum National d’Histoire Naturelle, Institut de Systématique, Evolution, Biodiversité (UMR 7205), Paris, France. [email protected] ABSTRACT A key is presented to seven genera and fifteen species of hornworts recorded from Colombia. Three species found in Ecuador but not yet in Colombia (Dendroceros crispatus, Phaeomegaceros squamuligerus, and Phaeoceros tenuis) are also included in the key. Key words. Biodiversity, identification, taxonomy. RESUMEN Se presenta una clave taxonómica para los siete géneros y quince especies de antocerotes registrados en Colombia. Tres especies registradas en Ecuador, pero aún no en Colombia (Dendroceros crispatus, Phaeomegaceros squamuligerus y Phaeoceros tenuis), también son incluidas. Palabras clave. Biodiversidad, identificación, taxonomía. INTRODUCCIÓN visible as black dots, rarely as blue lines (in Leiosporoceros); chloroplasts large, Hornworts (Anthocerotophyta) are a small 1–2(–4) per cell, frequently with a pyrenoid; division of bryophytes containing about 192 2) gametangia immersed in the thallus, accepted species worldwide (excluding 28 originating from an inner thallus cell; 3) doubtful species), in five families and 12 sporophyte narrowly cylindrical, without genera (Villarreal and Cargill 2016). They seta; 4) sporophyte growth by means of are commonly found on soil in rather open a basal meristem; 5) spore maturation places, but also on rotten logs, rock, bark asynchronous; and 6) capsule dehiscence or on living leaves. Hornworts were in the gradual, from the apex slowly downwards, past often classified with the liverworts by means of 2(-4) valves, rarely by an because of their superficial resemblance to operculum. -

A Revision of the Genus Anthoceros (Anthocerotaceae, Anthocerotophyta) in China
TERMS OF USE This pdf is provided by Magnolia Press for private/research use. Commercial sale or deposition in a public library or website is prohibited. Phytotaxa 100 (1): 21–35 (2013) ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ PHYTOTAXA Copyright © 2013 Magnolia Press Article ISSN 1179-3163 (online edition) http://dx.doi.org/10.11646/phytotaxa.100.1.3 A revision of the genus Anthoceros (Anthocerotaceae, Anthocerotophyta) in China TAO PENG1,2 & RUI-LIANG ZHU1* 1 Department of Biology, School of Life Science, East China Normal University, 3663 Zhong Shan North Road, Shanghai 200062, China; *Corresponding author: [email protected] 2 School of Life Science, Guizhou Normal University, 116 Bao Shan North Road, Guiyang 550001, China; [email protected] Abstract The genus Anthoceros (Anthocerotaceae, Anthocerotopsida) in China is reviewed. Five species and one variety are recognized. Anthoceros alpinus, A. bharadwajii, and A. subtilis, are reported new to China. Aspiromitus areolatus and Anthoceros esquirolii are proposed as new synonyms of Folioceros fuciformis and Phaeoceros carolinianus, respectively. A key to the species of Anthoceros in China is provided. Key words: Anthoceros alpinus, A. bharadwajii, A. subtilis, hornworts, new synonym Introduction Hornworts (Anthocerotophyta) represent a key group in the understanding of evolution of plant form because they are hypothesized to be sister to the tracheophytes (Qiu et al. 2006). An estimate of 200–250 species of hornworts exist worldwide (Villarreal et al. 2010; Garcia et al. 2012; Villarreal et al. 2012). Anthoceros Linnaeus (1753: 1139) is the largest genus of hornworts, with ca. 83 species (Villarreal et al. 2010). With a global distribution, the centres of diversity in the genus are in the Neotropics and tropical Africa and Asia. -

Print This Article
Annals of Plant Sciences 6.11 (2017) pp. 1758-1762 Research Article Enumeration of the new Hornworts from Bilaspur (Chhattisgarh), India. Mery Aradhna Kerketta and A.K. Dixit* Department of Botany, Guru Ghasidas Vishwavidyalaya, Koni, Bilaspur (Chhattisgarh), 495009, India. Received: 9/29/2017; Accepted: 10/15/2017 Abstract: A preliminary survey of Bilaspur-Achanakmar Tiger Reserve (ATR) forest area shows that phylogenetically significant hornworts are quite dominant in the area. The smallest known group of bryophytes Anthoceros (Hornworts) is a terrestrial and cosmopolitan genus characterized by horn-shaped sporophyte. Present investigation deals with the morphotaxonomic account of three species of hornworts. Notothylas levieri Schiffn. Ex Steph., Anthoceros punctatus L., Sp. and Phaeoceros leavies (L.) Prosk., has been identified from different localities of Bilaspur- Achnakmar Tigar Reserve (ATR) and Achanakmar–Amarkantak Biosphere Reserve (AABR), Chhattisgarh. The comprehensive and consolidate account, has been provided along with identification key. All three were new reports to the Chhattisgarh Bryoflora. Keywords: Hornworts, Bilaspur (AABR), Anthoceroteceae, Notothylas, Morphotaxonomy Himalayas, Central India (Pachmarhi) and Introduction Uttarakhnad extended to Gangetic planes, whereas Bryophytes have a great diversity which includes Phaeoceros with four taxa is widely known in Western liverworts, Mosses and Hornworts. Bryophyte in and Eastern Himalayas in India and extended up to general Anthrocopsida includes six genera, all very arid regions of Rajasthan was reported by genera are usually placed in Anthocerotaceae. Srivastava 1998. During present investigation of Initially Muller (1941) recognize two family three new species were recorded from Bilaspur – Anthocerotaceae and Notothylaceae with single ATR and AABR regions, these three identified new genus Notothylas Sull. -

Anthoceros Genomes Illuminate the Origin of Land Plants and the Unique Biology of Hornworts
ARTICLES https://doi.org/10.1038/s41477-020-0618-2 Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts Fay-Wei Li 1,2 ✉ , Tomoaki Nishiyama 3, Manuel Waller4, Eftychios Frangedakis5, Jean Keller 6, Zheng Li7, Noe Fernandez-Pozo 8, Michael S. Barker 7, Tom Bennett 9, Miguel A. Blázquez 10, Shifeng Cheng11, Andrew C. Cuming 9, Jan de Vries 12, Sophie de Vries 13, Pierre-Marc Delaux 6, Issa S. Diop4, C. Jill Harrison14, Duncan Hauser1, Jorge Hernández-García 10, Alexander Kirbis4, John C. Meeks15, Isabel Monte 16, Sumanth K. Mutte 17, Anna Neubauer4, Dietmar Quandt18, Tanner Robison1,2, Masaki Shimamura19, Stefan A. Rensing 8,20,21, Juan Carlos Villarreal 22,23, Dolf Weijers 17, Susann Wicke 24, Gane K.-S. Wong 25,26, Keiko Sakakibara 27 and Péter Szövényi 4,28 ✉ Hornworts comprise a bryophyte lineage that diverged from other extant land plants >400 million years ago and bears unique biological features, including a distinct sporophyte architecture, cyanobacterial symbiosis and a pyrenoid-based carbon- concentrating mechanism (CCM). Here, we provide three high-quality genomes of Anthoceros hornworts. Phylogenomic analy- ses place hornworts as a sister clade to liverworts plus mosses with high support. The Anthoceros genomes lack repeat-dense centromeres as well as whole-genome duplication, and contain a limited transcription factor repertoire. Several genes involved in angiosperm meristem and stomatal function are conserved in Anthoceros and upregulated during sporophyte development, suggesting possible homologies at the genetic level. We identified candidate genes involved in cyanobacterial symbiosis and found that LCIB, a Chlamydomonas CCM gene, is present in hornworts but absent in other plant lineages, implying a possible conserved role in CCM function. -

On Anthoceros Phymatodes M. Howe and the Hornwort Genus Phymatoceros Stotler, W. T. Doyle & Crand.-Stotl. (Anthocerotophyta)
Cryptogamie,Bryologie,2006,27 (1):59-73 ©2006 Adac.Tous droits réservés On Anthoceros phymatodes M.Howe and the hornwort genus Phymatoceros Stotler, W.T.Doyle & Crand.-Stotl. (Anthocerotophyta) 1 BarbaraJ.CRANDALL-STOTLER a,*,Raymond E.STOTLER a & WilliamT.DOYLE b a Department of Plant Biology,Southern Illinois University, Carbondale,IL 62901-6509,U.S.A. b Department of Ecology and Evolutionary Biology, University of California, Santa Cruz,CA 95064,U.S.A. (Received 20 September 2005,accepted 28 November 2005) Abstract – Based upon our study of type specimens,we confirm that Anthoceros phyma- todes M.Howe from California is synonymous with Phaeoceros bulbiculosus (Brot.) Prosk. from Portugal. Furthermore,this taxon forms the basis for the recently named,monotypic genus Phymatoceros Stotler,W.T.Doyle & Crand.-Stotl. Our observations of the mor- phology,anatomy,and phenology of living populations from both California and Portugal reveal a suite of characters that discriminate this taxon,not only from Phaeoceros Prosk., but also from all other hornwort genera.These include rounded to spindle-shaped chloro- plasts with abundant,bulging starch grains that may obscure a pyrenoid;highly dimorphic, dioicous thalli;a single antheridium per antheridial chamber,rather than 2 to 4 as in Phaeoceros ;and spores that are fuscous at maturity. Anthocerotophyta / California / Hornworts / Mediterranean / Phaeoceros / Phymatoceros / tubers INTRODUCTION Marshall Howe collected a hornwort on March 19,1892 near “The Old Mill” in Marin County,California that although lacking reproductive structures, was yet of interest because it bore stalked ventral tubers.These tubers are initi- ated at the growing point apex and become ventral in the central midrib region with subsequent thallus growth (vide Howe,1898).