A Simple Agrobacterium-Mediated Stable Transformation Technique for the Hornwort Model Anthoceros Agrestis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novelties in the Hornwort Flora of Croatia and Southeast Europe
cryptogamie Bryologie 2019 ● 40 ● 22 DIRECTEUR DE LA PUBLICATION : Bruno David, Président du Muséum national d’Histoire naturelle RÉDACTEURS EN CHEF / EDITORS-IN-CHIEF : Denis LAMY ASSISTANTS DE RÉDACTION / ASSISTANT EDITORS : Marianne SALAÜN ([email protected]) MISE EN PAGE / PAGE LAYOUT : Marianne SALAÜN RÉDACTEURS ASSOCIÉS / ASSOCIATE EDITORS Biologie moléculaire et phylogénie / Molecular biology and phylogeny Bernard GOFFINET Department of Ecology and Evolutionary Biology, University of Connecticut (United States) Mousses d’Europe / European mosses Isabel DRAPER Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) Francisco LARA GARCÍA Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) Mousses d’Afrique et d’Antarctique / African and Antarctic mosses Rysiek OCHYRA Laboratory of Bryology, Institute of Botany, Polish Academy of Sciences, Krakow (Pologne) Bryophytes d’Asie / Asian bryophytes Rui-Liang ZHU School of Life Science, East China Normal University, Shanghai (China) Bioindication / Biomonitoring Franck-Olivier DENAYER Faculté des Sciences Pharmaceutiques et Biologiques de Lille, Laboratoire de Botanique et de Cryptogamie, Lille (France) Écologie des bryophytes / Ecology of bryophyte Nagore GARCÍA MEDINA Department of Biology (Botany), and Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) COUVERTURE / COVER : Extraits d’éléments de la Figure 2 / Extracts of -

Phytotaxa, a Synthesis of Hornwort Diversity
Phytotaxa 9: 150–166 (2010) ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ Article PHYTOTAXA Copyright © 2010 • Magnolia Press ISSN 1179-3163 (online edition) A synthesis of hornwort diversity: Patterns, causes and future work JUAN CARLOS VILLARREAL1 , D. CHRISTINE CARGILL2 , ANDERS HAGBORG3 , LARS SÖDERSTRÖM4 & KAREN SUE RENZAGLIA5 1Department of Ecology and Evolutionary Biology, University of Connecticut, 75 North Eagleville Road, Storrs, CT 06269; [email protected] 2Centre for Plant Biodiversity Research, Australian National Herbarium, Australian National Botanic Gardens, GPO Box 1777, Canberra. ACT 2601, Australia; [email protected] 3Department of Botany, The Field Museum, 1400 South Lake Shore Drive, Chicago, IL 60605-2496; [email protected] 4Department of Biology, Norwegian University of Science and Technology, N-7491 Trondheim, Norway; [email protected] 5Department of Plant Biology, Southern Illinois University, Carbondale, IL 62901; [email protected] Abstract Hornworts are the least species-rich bryophyte group, with around 200–250 species worldwide. Despite their low species numbers, hornworts represent a key group for understanding the evolution of plant form because the best–sampled current phylogenies place them as sister to the tracheophytes. Despite their low taxonomic diversity, the group has not been monographed worldwide. There are few well-documented hornwort floras for temperate or tropical areas. Moreover, no species level phylogenies or population studies are available for hornworts. Here we aim at filling some important gaps in hornwort biology and biodiversity. We provide estimates of hornwort species richness worldwide, identifying centers of diversity. We also present two examples of the impact of recent work in elucidating the composition and circumscription of the genera Megaceros and Nothoceros. -

Anthocerotophyta
Glime, J. M. 2017. Anthocerotophyta. Chapt. 2-8. In: Glime, J. M. Bryophyte Ecology. Volume 1. Physiological Ecology. Ebook 2-8-1 sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 5 June 2020 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 2-8 ANTHOCEROTOPHYTA TABLE OF CONTENTS Anthocerotophyta ......................................................................................................................................... 2-8-2 Summary .................................................................................................................................................... 2-8-10 Acknowledgments ...................................................................................................................................... 2-8-10 Literature Cited .......................................................................................................................................... 2-8-10 2-8-2 Chapter 2-8: Anthocerotophyta CHAPTER 2-8 ANTHOCEROTOPHYTA Figure 1. Notothylas orbicularis thallus with involucres. Photo by Michael Lüth, with permission. Anthocerotophyta These plants, once placed among the bryophytes in the families. The second class is Leiosporocerotopsida, a Anthocerotae, now generally placed in the phylum class with one order, one family, and one genus. The genus Anthocerotophyta (hornworts, Figure 1), seem more Leiosporoceros differs from members of the class distantly related, and genetic evidence may even present -

Introduction to Common Native & Invasive Freshwater Plants in Alaska
Introduction to Common Native & Potential Invasive Freshwater Plants in Alaska Cover photographs by (top to bottom, left to right): Tara Chestnut/Hannah E. Anderson, Jamie Fenneman, Vanessa Morgan, Dana Visalli, Jamie Fenneman, Lynda K. Moore and Denny Lassuy. Introduction to Common Native & Potential Invasive Freshwater Plants in Alaska This document is based on An Aquatic Plant Identification Manual for Washington’s Freshwater Plants, which was modified with permission from the Washington State Department of Ecology, by the Center for Lakes and Reservoirs at Portland State University for Alaska Department of Fish and Game US Fish & Wildlife Service - Coastal Program US Fish & Wildlife Service - Aquatic Invasive Species Program December 2009 TABLE OF CONTENTS TABLE OF CONTENTS Acknowledgments ............................................................................ x Introduction Overview ............................................................................. xvi How to Use This Manual .................................................... xvi Categories of Special Interest Imperiled, Rare and Uncommon Aquatic Species ..................... xx Indigenous Peoples Use of Aquatic Plants .............................. xxi Invasive Aquatic Plants Impacts ................................................................................. xxi Vectors ................................................................................. xxii Prevention Tips .................................................... xxii Early Detection and Reporting -

Aquatic Vegetation Control in Arkansas George Selden, Extension Aquaculture Specialist
MP556 Aquatic Vegetation Control in Arkansas George Selden, Extension Aquaculture Specialist University of Arkansas at Pine Blu, United States Department of Agriculture, and County Governments Cooperating TABLE OF CONTENTS Introduction...............................................................................................................................................2 Aquatic Plant Identification.....................................................................................................................2 Control Techniques...................................................................................................................................3 Herbicide Selection..................................................................................................................................6 Herbicide Types.........................................................................................................................................6 Why Treatments Fail.................................................................................................................................7 Herbicide Formulations............................................................................................................................7 Herbicide Application and Application Equipment............................................................................12 Herbicide Application Rate Calculation and Pond Size Determination..........................................14 Aquatic Plants that Commonly Become Problems -

Morphology, Anatomy and Reproduction of Anthoceros
Morphology, Anatomy and Reproduction of Anthoceros - INDRESH KUMAR PANDEY Taxonomic Position of Anthoceros Class- Anthocerotopsida Single order Anthocerotales Two families Anthocerotaceae Notothylaceae Representative Genus Anthoceros Notothylus General features of Anthocerotopsida • Forms an isolated evolutionary line • Sometimes considered independent from Bryophytes and placed in division Anthocerophyta • Called as Hornworts due to horn like structure of sporophyte • Commonly recognised genera includes Anthoceros, Megaceros, Nothothylus, Dendroceros Anthoceros :Habitat & Distribution • Cosmopolitan • Mainly in temperate & tropical regions • More than 200 species, 25 sp. Recorded from India. • Mostly grows in moist shady places, sides of ditches or in moist hollows among rocks • Few species grow on decaying wood. • Three common Indian species- A. erectus, A. crispulus, A. himalayensis Anthoceros: Morphology Dorsal surface Ventral surface Rhizoids (smooth walled) Thallus showing tubers External features • Thallus (gametophyte)- small, dark green, dorsiventral, prostrate, branched or lobed • No midrib, spongy due to presence of underlying mucilaginous ducts • Dorsal surface varies from species to species Smooth- A. laevis Velvety- A. crispulus Rough- A. fusiformis • Smooth walled rhizoid on ventral surface • Rounded bluish green thickened area on ventral surface- Nostoc colonies Internal structure Vertical Transverse Section- Diagrammatic Vertical Transverse Section- Cellular Internal Structure • Simple, without cellular differentiation -

Fuller’S Leadership and Over- Vincent of the Refuge Staff Are Notable for Having Sight Were Invaluable
Acknowledgments Acknowledgments Many people have contributed to this plan over many detailed and technical requirements of sub- the last seven years. Several key staff positions, missions to the Service, the Environmental Protec- including mine, have been filled by different people tion Agency, and the Federal Register. Jon during the planning period. Tom Palmer and Neil Kauffeld’s and Nita Fuller’s leadership and over- Vincent of the Refuge staff are notable for having sight were invaluable. We benefited from close col- been active in the planning for the entire extent. laboration and cooperation with staff of the Illinois Tom and Neil kept the details straight and the rest Department of Natural Resources. Their staff par- of us on track throughout. Mike Brown joined the ticipated from the early days of scoping through staff in the midst of the process and contributed new reviews and re-writes. We appreciate their persis- insights, analysis, and enthusiasm that kept us mov- tence, professional expertise, and commitment to ing forward. Beth Kerley and John Magera pro- our natural resources. Finally, we value the tremen- vided valuable input on the industrial and public use dous involvement of citizens throughout the plan- aspects of the plan. Although this is a refuge plan, ning process. We heard from visitors to the Refuge we received notable support from our regional office and from people who care about the Refuge without planning staff. John Schomaker provided excep- ever having visited. Their input demonstrated a tional service coordinating among the multiple level of caring and thought that constantly interests and requirements within the Service. -

Establishment of Anthoceros Agrestis As a Model Species for Studying the Biology of Hornworts
Szövényi et al. BMC Plant Biology (2015) 15:98 DOI 10.1186/s12870-015-0481-x METHODOLOGY ARTICLE Open Access Establishment of Anthoceros agrestis as a model species for studying the biology of hornworts Péter Szövényi1,2,3,4†, Eftychios Frangedakis5,8†, Mariana Ricca1,3, Dietmar Quandt6, Susann Wicke6,7 and Jane A Langdale5* Abstract Background: Plants colonized terrestrial environments approximately 480 million years ago and have contributed significantly to the diversification of life on Earth. Phylogenetic analyses position a subset of charophyte algae as the sister group to land plants, and distinguish two land plant groups that diverged around 450 million years ago – the bryophytes and the vascular plants. Relationships between liverworts, mosses hornworts and vascular plants have proven difficult to resolve, and as such it is not clear which bryophyte lineage is the sister group to all other land plants and which is the sister to vascular plants. The lack of comparative molecular studies in representatives of all three lineages exacerbates this uncertainty. Such comparisons can be made between mosses and liverworts because representative model organisms are well established in these two bryophyte lineages. To date, however, a model hornwort species has not been available. Results: Here we report the establishment of Anthoceros agrestis as a model hornwort species for laboratory experiments. Axenic culture conditions for maintenance and vegetative propagation have been determined, and treatments for the induction of sexual reproduction and sporophyte development have been established. In addition, protocols have been developed for the extraction of DNA and RNA that is of a quality suitable for molecular analyses. -

About the Book the Format Acknowledgments
About the Book For more than ten years I have been working on a book on bryophyte ecology and was joined by Heinjo During, who has been very helpful in critiquing multiple versions of the chapters. But as the book progressed, the field of bryophyte ecology progressed faster. No chapter ever seemed to stay finished, hence the decision to publish online. Furthermore, rather than being a textbook, it is evolving into an encyclopedia that would be at least three volumes. Having reached the age when I could retire whenever I wanted to, I no longer needed be so concerned with the publish or perish paradigm. In keeping with the sharing nature of bryologists, and the need to educate the non-bryologists about the nature and role of bryophytes in the ecosystem, it seemed my personal goals could best be accomplished by publishing online. This has several advantages for me. I can choose the format I want, I can include lots of color images, and I can post chapters or parts of chapters as I complete them and update later if I find it important. Throughout the book I have posed questions. I have even attempt to offer hypotheses for many of these. It is my hope that these questions and hypotheses will inspire students of all ages to attempt to answer these. Some are simple and could even be done by elementary school children. Others are suitable for undergraduate projects. And some will take lifelong work or a large team of researchers around the world. Have fun with them! The Format The decision to publish Bryophyte Ecology as an ebook occurred after I had a publisher, and I am sure I have not thought of all the complexities of publishing as I complete things, rather than in the order of the planned organization. -

Hornwort Pyrenoids, Carbon-Concentrating Structures, Evolved and Were Lost at Least five Times During the Last 100 Million Years
Hornwort pyrenoids, carbon-concentrating structures, evolved and were lost at least five times during the last 100 million years Juan Carlos Villarreal1 and Susanne S. Renner Systematic Botany and Mycology, Department of Biology, University of Munich (LMU), Munich 80638, Germany Edited by John Raven, University of Dundee, Dundee, United Kingdom, and accepted by the Editorial Board September 24, 2012 (received for review August 7, 2012) Ribulose-1,5-Biphosphate-carboxylase-oxygenase (RuBisCO) has a have a stacked arrangement of thylakoid membranes (grana) that crucial role in carbon fixation but a slow catalytic rate, a problem results in the spatial separation of photosystems and increases the overcome in some plant lineages by physiological and anatomical efficiency of light capture in terrestrial environments (13). Horn- traits that elevate carbon concentrations around the enzyme. Such wort grana consist of stacks of short thylakoids and lack end carbon-concentrating mechanisms are hypothesized to have evolved membranes. Therefore, unlike other land plants, hornwort grana during periods of low atmospheric CO2. Hornworts, the sister to are devoid of the membrane “sacs” that enclose intrathylakoid vascular plants, have a carbon-concentrating mechanism that relies spaces. Presumably, the perpendicular channel thylakoid system in on pyrenoids, proteinaceous bodies mostly consisting of RuBisCO. hornwort plastids serves to isolate biochemical processes (13). We generated a phylogeny based on mitochondrial and plastid Organic isotope discrimination supports a function in CO2 sequences for 36% of the approximately 200 hornwort species to concentration for hornwort pyrenoids (14–18). Mass spectrometry infer the history of gains and losses of pyrenoids in this clade; we analyses show that hornworts with pyrenoids (e.g., Phaeoceros and also used fossils and multiple dating approaches to generate a chro- Notothylas) have lower compensation points (11–13 vs. -
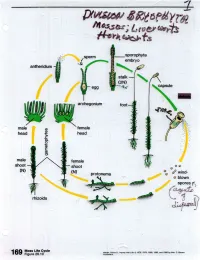
02-Bryophyta-2.Pdf
498 INTRODUCTORY PLANT SCIENCE layer. In certain species, the capsule con til1Ues to grow as long as the gametophyte lives. The presence of a meristcm in A11tlioceros and an aerating system complete with sto mata may indicate that Antlwceros evolved from ancestors with even ·larger· and more 1. Civ complex sporophytes. These features may Bryophyt be vestiges from a more complex ancestral. 2. Wh sporophyte. that ti alga ? 3. De� ORIGIN AND RELATIONSHIPS OF THE Bryophyt, BRYOPHYTA 4._ characteri Little is known with certainty about the r are small origin and evolution of the bryophytes. The ' tall. ( B) fossil record is too fragmenta to enable • ry s true roots us to trace their evolutionary history. Frag ,, sperms ar mentary remnants of thallose liverworts, spore idium and mother\4'/ttN) which resemble present-day liverworts, have cell Jar arch� been foum! in rocks of Carboniferous age, bryophyt as have structures that may be remains of more com mosses. all bryoph The immediate ancestors of the bryo bryo whicl phytes were probably more complex plants from the have an i than present-day forms. In other words, the gametophy evolutionary tendency has been one of re- with a dip1 duction instead of increased complexity. •• • • • spores If evolution has progressed . from more • . •, .... • s. __ complex sporophytes to those of simpler • . vascular p Anthoceros . form, the sporophyte of wou1d • • green algat be considered more ancient than that of • (D) red Marchantia or Riccia. Because Riccia has Fig. 35-16. longitudinal section of the spo 6. How the most reduced sporophyte, it would be t·:i;,hyte of Anthoceros. -

Laurentian-Acadian Alkaline Conifer-Hardwood Swamp
Laurentian-Acadian Alkaline Conifer-Hardwood Swamp Macrogroup: Northern Swamp yourStateNatural Heritage Ecologist for more information about this habitat. This is modeledmap a distributiononbased current and is data nota substitute for field inventory. based Contact © Elizabeth Thompson (Vermont Land Trust) Description: A forested swamp of alkaline wetlands associated with limestone or other calcareous substrate in the northern part of the glaciated northeast. Northern white cedar is often present and may dominate the canopy or be mixed with other conifers or with deciduous trees, most commonly red maple or black ash. Some examples can be almost entirely deciduous and dominated by black ash. Red-osier dogwood is a common shrub. The herb layer tends to be more diverse than in acidic swamps, due to higher pH and nutrient level. Small open fenny areas may occur within the wetland. The moss layer is often extensive and diverse. Seepage may influence parts of the wetland, but the hydrology is State Distribution: CT, MA, ME, NH, NY, VT dominated by the basin setting. Total Habitat Acreage: 921,478 Ecological Setting and Natural Processes: Percent Conserved: 19.5% These forested wetlands are uncommon in the glaciated State State GAP 1&2 GAP 3 Unsecured northeast except in areas with extensive limestone or similar State Habitat % Acreage (acres) (acres) (acres) substrate. The substrate is typically mineral soil, but there ME 56% 520,121 14,203 60,307 445,611 may be some peat, and there is often direct contact with NY 38% 345,750 49,536 44,764 251,450 alkaline groundwater. VT 5% 43,899 1,177 4,786 37,935 NH 1% 7,363 2,054 1,013 4,295 MA 0% 4,261 643 1,267 2,350 CT 0% 86 0 0 86 Similar Habitat Types: Similar to North-Central Interior and Appalachian Rich Swamp, but with a flora characteristic of a cooler climate.