Computational Drug Target Screening Through Protein Interaction Profiles
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Information
Supplementary Information Network-based Drug Repurposing for Novel Coronavirus 2019-nCoV Yadi Zhou1,#, Yuan Hou1,#, Jiayu Shen1, Yin Huang1, William Martin1, Feixiong Cheng1-3,* 1Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA 2Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH 44195, USA 3Case Comprehensive Cancer Center, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA #Equal contribution *Correspondence to: Feixiong Cheng, PhD Lerner Research Institute Cleveland Clinic Tel: +1-216-444-7654; Fax: +1-216-636-0009 Email: [email protected] Supplementary Table S1. Genome information of 15 coronaviruses used for phylogenetic analyses. Supplementary Table S2. Protein sequence identities across 5 protein regions in 15 coronaviruses. Supplementary Table S3. HCoV-associated host proteins with references. Supplementary Table S4. Repurposable drugs predicted by network-based approaches. Supplementary Table S5. Network proximity results for 2,938 drugs against pan-human coronavirus (CoV) and individual CoVs. Supplementary Table S6. Network-predicted drug combinations for all the drug pairs from the top 16 high-confidence repurposable drugs. 1 Supplementary Table S1. Genome information of 15 coronaviruses used for phylogenetic analyses. GenBank ID Coronavirus Identity % Host Location discovered MN908947 2019-nCoV[Wuhan-Hu-1] 100 Human China MN938384 2019-nCoV[HKU-SZ-002a] 99.99 Human China MN975262 -

Pharmaceuticals As Environmental Contaminants
PharmaceuticalsPharmaceuticals asas EnvironmentalEnvironmental Contaminants:Contaminants: anan OverviewOverview ofof thethe ScienceScience Christian G. Daughton, Ph.D. Chief, Environmental Chemistry Branch Environmental Sciences Division National Exposure Research Laboratory Office of Research and Development Environmental Protection Agency Las Vegas, Nevada 89119 [email protected] Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada Why and how do drugs contaminate the environment? What might it all mean? How do we prevent it? Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada This talk presents only a cursory overview of some of the many science issues surrounding the topic of pharmaceuticals as environmental contaminants Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada A Clarification We sometimes loosely (but incorrectly) refer to drugs, medicines, medications, or pharmaceuticals as being the substances that contaminant the environment. The actual environmental contaminants, however, are the active pharmaceutical ingredients – APIs. These terms are all often used interchangeably Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada Office of Research and Development Available: http://www.epa.gov/nerlesd1/chemistry/pharma/image/drawing.pdfNational -

Interaction of Benzodiazepines with Central Nervous Glycine Receptors: Possible Mechanism of Action (Strychnine Binding/Behavioral Tests) ANNE B
Proc. Nat. Acad. Sci. USA Vol. 71, No. 6, pp. 2246-2250, June 1974 Interaction of Benzodiazepines with Central Nervous Glycine Receptors: Possible Mechanism of Action (strychnine binding/behavioral tests) ANNE B. YOUNG, STEPHEN R. ZUKIN, AND SOLOMON H. SNYDER* Departments of Pharmacology and Experimental Therapeutics and Psychiatry and the Behavioral Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205 Communicaed by Cheves Walling, January 29, 1974 ABSTRACT Interaction of 21 benzodiazepines with the ously (4). The specific activity of the labeled strychnine was glycine receptor in the brainstem and spinal cord of rat have been evaluated in terms of their displacement of 13 Ci/mmol, with concentrations determined by comparison ['HIstrychnine binding. The rank order of potency of the with the ultraviolet absorption of standard solutions at 254 21 drugs in displacing specific ['H1strychnine binding cor- nm. relates (p < 0.005) with their rank order of potency in a variety of pharmacological and behavioral tests in Tissue Preparation. Male Sprague-Dawley rats (100-150 g) humans and animals that predict clinical efficacy. There were decapitated and the brainstems and spinal cords re- is a 50-fold variation in potency of the series of benzo- moved. Crude synaptic membrane fractions of these tissues diazepines with mean effective dose (ED5.) values ranging were prepared by the differential centrifugation technique from 19juM to >1000 pM. Diazepam (Valium®) and chlor- diazepoxide (Librium®) have EDN's of 26 MM and 200 pM, previously reported (4). respectively, whereas the ED5. for glycine is 25 ;M. The in- hibitory effects of 10 of the agents in two other central ner- Binding A8say. -

Therapeutic Applications of the Carbonic Anhydrase Inhibitors
REVIEW Therapeutic applications of the carbonic anhydrase inhibitors Claudiu T Supuran Inhibitors of the zinc enzyme carbonic anhydrase (CA) targeting different isozymes among Università degli Studi, Laboratorio di Chimica the 15 presently reported in humans have various therapeutic applications. These enzymes Bioinorganica, Rm 188, are efficient catalysts for the hydration of carbon dioxide to bicarbonate and protons, Via della Lastruccia 3, playing crucial physiological/pathological roles related to acid–base homeostasis, secretion I-50019 Sesto Fiorentino (Firenze), Italy of electrolytes, transport of ions, biosynthetic reactions and tumorigenesis. Many CA Tel.: +39 055 457 3005; inhibitors have been reported as antiglaucoma, anticancer and antiobesity agents, or for Fax: +39 055 457 3385; the management of a variety of neurological disorders, including epilepsy and altitude E-mail: claudiu.supuran@ unifi.it disease. Furthermore, some CA isozymes appear to be valuable markers of tumor progression in a variety of tissues/organs. Various CAs present in pathogens such as Plasmodium falciparum, Helicobacter pylori, Mycobacterium tuberculosis, Candida albicans and Cryptococcus neoformans, have started to also be investigated recently as drug targets. Recent progress in all these areas, as well as in the characterization of new such enzymes isolated in many other organisms/tissues, in addition to their detailed catalytic/inhibition mechanisms, are reviewed herein, together with the therapeutic use of these enzyme inhibitors and drug-design studies for obtaining such new-generation derivatives with improved activity. Among the zinc enzymes extensively studied, forms (CA VA and CA VB), as well as a the carbonic anhydrases (CAs) occupy a special secreted CA isozyme (in saliva and milk; place for several reasons. -

Identifying Nootropic Drug Targets Via Large-Scale Cognitive GWAS and Transcriptomics
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.06.934752; this version posted February 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: Identifying Nootropic Drug Targets via Large-Scale Cognitive GWAS and Transcriptomics Max Lam1, 2, 3,4, Chia-Yen, Chen3,5,6, Xia Yan7,8, W. David Hill9, 10, Joey W. Trampush11, Jin Yu1, Emma Knowles12,13,14, Gail Davies9, 10, Eli Stahl15, 16, Laura Huckins15, 16, David C. Liewald10, Srdjan Djurovic17, 18, Ingrid Melle18, 19, Andrea Christoforou20, Ivar Reinvang21, Pamela DeRosse1, 22, 23, Astri J. Lundervold24, Vidar M. Steen18, 20, Thomas Espeseth19, 21, Katri Räikkönen25, Elisabeth Widen26, Aarno Palotie26, 27, 28, Johan G. Eriksson29, 30, 31, Ina Giegling32, Bettina Konte32, Annette M. Hartmann32, Panos Roussos15, 16, 33, Stella Giakoumaki34, Katherine E. Burdick15, 33, 35, Antony Payton36, William Ollier37, 38, Ornit Chiba- Falek39, Deborah K. Koltai39, 40 , Anna C. Need41, Elizabeth T. Cirulli42, Aristotle N. Voineskos43, Nikos C. Stefanis44, 45, 46, Dimitrios Avramopoulos47, 48, Alex Hatzimanolis44, 45, 46, Nikolaos Smyrnis44, 45, Robert M. Bilder49, Nelson A. Freimer49, Tyrone D. Cannon50, 51, Edythe London49, Russell A. Poldrack52, Fred W. Sabb53, Eliza Congdon49, Emily Drabant Conley54, Matthew A. Scult55, Dwight Dickinson56, Richard E. Straub57, Gary Donohoe58, Derek Morris58, Aiden Corvin59, Michael Gill59, Ahmad R. Hariri55, Daniel R. Weinberger57, Neil Pendleton60, Panos Bitsios61, Dan Rujescu32, Jari Lahti25, 62, Stephanie Le Hellard18, 20, Matthew C. -

Wo 2008/127291 A2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (43) International Publication Date PCT (10) International Publication Number 23 October 2008 (23.10.2008) WO 2008/127291 A2 (51) International Patent Classification: Jeffrey, J. [US/US]; 106 Glenview Drive, Los Alamos, GOlN 33/53 (2006.01) GOlN 33/68 (2006.01) NM 87544 (US). HARRIS, Michael, N. [US/US]; 295 GOlN 21/76 (2006.01) GOlN 23/223 (2006.01) Kilby Avenue, Los Alamos, NM 87544 (US). BURRELL, Anthony, K. [NZ/US]; 2431 Canyon Glen, Los Alamos, (21) International Application Number: NM 87544 (US). PCT/US2007/021888 (74) Agents: COTTRELL, Bruce, H. et al.; Los Alamos (22) International Filing Date: 10 October 2007 (10.10.2007) National Laboratory, LGTP, MS A187, Los Alamos, NM 87545 (US). (25) Filing Language: English (81) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of national protection available): AE, AG, AL, AM, AT,AU, AZ, BA, BB, BG, BH, BR, BW, BY,BZ, CA, CH, (30) Priority Data: CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, 60/850,594 10 October 2006 (10.10.2006) US ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, KZ, LA, LC, LK, (71) Applicants (for all designated States except US): LOS LR, LS, LT, LU, LY,MA, MD, ME, MG, MK, MN, MW, ALAMOS NATIONAL SECURITY,LLC [US/US]; Los MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PG, PH, PL, Alamos National Laboratory, Lc/ip, Ms A187, Los Alamos, PT, RO, RS, RU, SC, SD, SE, SG, SK, SL, SM, SV, SY, NM 87545 (US). -

Analysis of US FDA-Approved Drugs Containing Sulfur Atoms
Top Curr Chem (Z) (2018) 376:5 https://doi.org/10.1007/s41061-018-0184-5 REVIEW Analysis of US FDA‑Approved Drugs Containing Sulfur Atoms Kevin A. Scott1,2 · Jon T. Njardarson1 Received: 20 November 2017 / Accepted: 5 January 2018 / Published online: 22 January 2018 © Springer International Publishing AG, part of Springer Nature 2018 Abstract In this review, we discuss all sulfur-containing FDA-approved drugs and their structures. The second section of the review is dedicated to structural analy- sis and is divided into 14 subsections, each focusing on one type of sulfur-contain- ing moiety. A concise graphical representation of each class features drugs that are organized on the basis of structural similarity, evolutionary relevance, and medical indication. This review ofers a unique and comprehensive overview of the struc- tural features of all sulfur-containing FDA-approved drugs to date. Keywords Sulfonamide · Thioether · Sulfoxide · Sulfone · Sulfate · Sulfur heterocycle Abbreviations 6-APA 6-Aminopenicillanic acid ADP Adenosine diphosphate cGMP Cyclic guanosine monophosphate COX-2 Cyclooxygenase-2 GERD Gastroesophageal refux disease GPCR G protein-coupled receptor HIV Human immunodefciency virus mRNA Messenger RNA NSAID Nonsteroidal anti-infammatory drugs SMN1 Survival motor neuron 1 This article is part of the Topical Collection “Sulfur Chemistry”, edited by Xuefeng Jiang. * Jon T. Njardarson [email protected] 1 Department of Chemistry and Biochemistry, University of Arizona, Tucson, AZ 85721, USA 2 Department of Pharmacology -

ARCI Uniform Classification Guidelines for Foreign Substances, Or Similar State Regulatory Guidelines, Shall Be Assigned Points As Follows
DRUG TESTING STANDARDS AND PRACTICES PROGRAM. Uniform Classification Guidelines for Foreign Substances And Recommended Penalties Model Rule. January, 2019 (V.14.0) © ASSOCIATION OF RACING COMMISSIONERS INTERNATIONAL – 2019. Association of Racing Commissioners International 2365 Harrodsburg Road- B450 Lexington, Kentucky, USA www.arci.com Page 1 of 66 Preamble to the Uniform Classification Guidelines of Foreign Substances The Preamble to the Uniform Classification Guidelines was approved by the RCI Drug Testing and Quality Assurance Program Committee (now the Drug Testing Standards and Practices Program Committee) on August 26, 1991. Minor revisions to the Preamble were made by the Drug Classification subcommittee (now the Veterinary Pharmacologists Subcommittee) on September 3, 1991. "The Uniform Classification Guidelines printed on the following pages are intended to assist stewards, hearing officers and racing commissioners in evaluating the seriousness of alleged violations of medication and prohibited substance rules in racing jurisdictions. Practicing equine veterinarians, state veterinarians, and equine pharmacologists are available and should be consulted to explain the pharmacological effects of the drugs listed in each class prior to any decisions with respect to penalities to be imposed. The ranking of drugs is based on their pharmacology, their ability to influence the outcome of a race, whether or not they have legitimate therapeutic uses in the racing horse, or other evidence that they may be used improperly. These classes of drugs are intended only as guidelines and should be employed only to assist persons adjudicating facts and opinions in understanding the seriousness of the alleged offenses. The facts of each case are always different and there may be mitigating circumstances which should always be considered. -

Is Based Partly on a Stored Dictionary and Partly on the Identification of Text
DOCUMENT RESUME ED 022 504 LI 000 909 By- Artandi, Susan; Baxenda!e, Stanley PROJECT MEDICO; MODEL EXPERIMENT IN DRUG INDEXING BY COMPUTER. FIRSTPROGRESS REPORT. Rutgers, The State Univ., New Brunswick, N.J. Graduate School of Library Service. Spons Agency-Public Health Service (DHEW), Washington, D.C. National Libraryof Medicine. Report No-PHS-LM-94 Pub Date Jan 68 Note- 1 1 1p. EDRS Price MF-$0.50 HC-1,452 Descriptors-*AUTOMATION, COMPUTER PROGRAMS, COMPUTERS, *COMPUTER STORAGE DEVICES, DICTIONARIES, *INDUING,*INFORMATIONRETRIEVAL,*INFORMATION STORAGE, INPUT OUTPUT, OPERATIONS RESEARCH, SEARCH STRATEGIES Identifiers- MEDIrCO, *Model Experiment in Drug Indexing by Computer This report describes Phase 1 of a research project with theover-all objective of exploring the applicability of automatic methods in theindexing of drug information appearing in English natural language text.Two major phases of the research have been completed:(1)development oftheautomaticindexing method andits implementation on the test documents and (2) creation of a machinesearchable file for the storage of the index records generated through automaticincicxing. Indexing is based partly on a stored dictionaryand partly on the identification of text characteristics which can be used to signal to the computerthe presence of information to be indexed. The characteristics of the machine file wereestablished with the desired search capabilities in mind. The file stores documentreferences with their associated index terms. In addition to assigning index terms todocuments the computer program also automatically assignsweights to the index terms to indicate their relative importance in the document 'description. The next two majorphases of the work will involve the design and implementation of the search programand the evaluation of the automatic indexing method. -
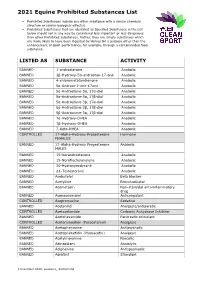
2021 Equine Prohibited Substances List
2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). Prohibited Substances that are identified as Specified Substances in the List below should not in any way be considered less important or less dangerous than other Prohibited Substances. Rather, they are simply substances which are more likely to have been ingested by Horses for a purpose other than the enhancement of sport performance, for example, through a contaminated food substance. LISTED AS SUBSTANCE ACTIVITY BANNED 1-androsterone Anabolic BANNED 3β-Hydroxy-5α-androstan-17-one Anabolic BANNED 4-chlorometatandienone Anabolic BANNED 5α-Androst-2-ene-17one Anabolic BANNED 5α-Androstane-3α, 17α-diol Anabolic BANNED 5α-Androstane-3α, 17β-diol Anabolic BANNED 5α-Androstane-3β, 17α-diol Anabolic BANNED 5α-Androstane-3β, 17β-diol Anabolic BANNED 5β-Androstane-3α, 17β-diol Anabolic BANNED 7α-Hydroxy-DHEA Anabolic BANNED 7β-Hydroxy-DHEA Anabolic BANNED 7-Keto-DHEA Anabolic CONTROLLED 17-Alpha-Hydroxy Progesterone Hormone FEMALES BANNED 17-Alpha-Hydroxy Progesterone Anabolic MALES BANNED 19-Norandrosterone Anabolic BANNED 19-Noretiocholanolone Anabolic BANNED 20-Hydroxyecdysone Anabolic BANNED Δ1-Testosterone Anabolic BANNED Acebutolol Beta blocker BANNED Acefylline Bronchodilator BANNED Acemetacin Non-steroidal anti-inflammatory drug BANNED Acenocoumarol Anticoagulant CONTROLLED Acepromazine Sedative BANNED Acetanilid Analgesic/antipyretic CONTROLLED Acetazolamide Carbonic Anhydrase Inhibitor BANNED Acetohexamide Pancreatic stimulant CONTROLLED Acetominophen (Paracetamol) Analgesic BANNED Acetophenazine Antipsychotic BANNED Acetophenetidin (Phenacetin) Analgesic BANNED Acetylmorphine Narcotic BANNED Adinazolam Anxiolytic BANNED Adiphenine Antispasmodic BANNED Adrafinil Stimulant 1 December 2020, Lausanne, Switzerland 2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). -

Drug/Substance Trade Name(S)
A B C D E F G H I J K 1 Drug/Substance Trade Name(s) Drug Class Existing Penalty Class Special Notation T1:Doping/Endangerment Level T2: Mismanagement Level Comments Methylenedioxypyrovalerone is a stimulant of the cathinone class which acts as a 3,4-methylenedioxypyprovaleroneMDPV, “bath salts” norepinephrine-dopamine reuptake inhibitor. It was first developed in the 1960s by a team at 1 A Yes A A 2 Boehringer Ingelheim. No 3 Alfentanil Alfenta Narcotic used to control pain and keep patients asleep during surgery. 1 A Yes A No A Aminoxafen, Aminorex is a weight loss stimulant drug. It was withdrawn from the market after it was found Aminorex Aminoxaphen, Apiquel, to cause pulmonary hypertension. 1 A Yes A A 4 McN-742, Menocil No Amphetamine is a potent central nervous system stimulant that is used in the treatment of Amphetamine Speed, Upper 1 A Yes A A 5 attention deficit hyperactivity disorder, narcolepsy, and obesity. No Anileridine is a synthetic analgesic drug and is a member of the piperidine class of analgesic Anileridine Leritine 1 A Yes A A 6 agents developed by Merck & Co. in the 1950s. No Dopamine promoter used to treat loss of muscle movement control caused by Parkinson's Apomorphine Apokyn, Ixense 1 A Yes A A 7 disease. No Recreational drug with euphoriant and stimulant properties. The effects produced by BZP are comparable to those produced by amphetamine. It is often claimed that BZP was originally Benzylpiperazine BZP 1 A Yes A A synthesized as a potential antihelminthic (anti-parasitic) agent for use in farm animals. -

Comprehensive Review of Visual Defects Reported with Topiramate
Journal name: Clinical Ophthalmology Article Designation: Review Year: 2017 Volume: 11 Clinical Ophthalmology Dovepress Running head verso: Ford et al Running head recto: Visual defects reported with topiramate open access to scientific and medical research DOI: http://dx.doi.org/10.2147/OPTH.S125768 Open Access Full Text Article REVIEW Comprehensive review of visual defects reported with topiramate Lisa Ford1 Objective: The objective of this study was to analyze clinical patterns of visual field defects Jeffrey L Goldberg2 (VFDs) reported with topiramate treatment and assess possible mechanism of action (MOA) Fred Selan1 for antiepileptic drug (AED) associated VFDs. Howard E Greenberg1 Methods: A comprehensive topiramate database review included preclinical data, sponsor’s Yingqi Shi1 clinical trials database, postmarketing spontaneous reports, and medical literature. All treatment- emergent adverse events (TEAEs) suggestive of retinal dysfunction/damage were summarized. 1Janssen Research & Development, Relative risk (RR) was computed from topiramate double-blind, placebo-controlled trials LLC, Titusville, NJ, 2Byers Eye Institute, Stanford University, (DBPCTs) data. Palo Alto, CA, USA Results: Preclinical studies and medical literature review suggested that despite sharing gamma-aminobutyric acid (GABA)-ergic MOA with other AEDs, topiramate treatment was not associated with VFDs. TEAEs suggestive of retinal dysfunction/damage were observed For personal use only. in 0.3%–0.7% of adults and pediatric patients with topiramate (N=4,679) versus #0.1% with placebo (N=1,834) in DBPCTs for approved indications (epilepsy and migraine prophylaxis); open-label trials (OLTs) and DBPCTs for investigational indications had similar incidence. Overall, 88% TEAEs were mild or moderate in severity. Serious TEAEs were very rare (DBPCTs: 0%; OLTs: #0.1%), and most were not treatment limiting, and resolved.