Handbook in Molecular Modeling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chem Compute Quickstart
Chem Compute Quickstart Chem Compute is maintained by Mark Perri at Sonoma State University and hosted on Jetstream at Indiana University. The Chem Compute URL is https://chemcompute.org/. We will use Chem Compute as the frontend for running electronic structure calculations with The General Atomic and Molecular Electronic Structure System, GAMESS (http://www.msg.ameslab.gov/gamess/). Chem Compute also provides access to other computational chemistry resources including PSI4 and the molecular dynamics packages TINKER and NAMD, though we will not be using those resource at this time. Follow this link, https://chemcompute.org/gamess/submit, to directly access the Chem Compute GAMESS guided submission interface. If you are a returning Chem Computer user, please log in now. If your University is part of the InCommon Federation you can log in without registering by clicking Login then "Log In with Google or your University" – select your University from the dropdown list. Otherwise if this is your first time working with Chem Compute, please register as a Chem Compute user by clicking the “Register” link in the top-right corner of the page. This gives you access to all of the computational resources available at ChemCompute.org and will allow you to maintain copies of your calculations in your user “Dashboard” that you can refer to later. Registering also helps track usage and obtain the resources needed to continue providing its service. When logged in with the GAMESS-Submit tabs selected, an instruction section appears on the left side of the page with instructions for several different kinds of calculations. -

Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department
Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department Name of FOSS Applications Employability GChemPaint GChemPaint is an editor for 2Dchem- GChemPaint is currently being developed ical structures with a multiple docu- as part of The Chemistry Development ment interface. Kit, and a Standard Widget Tool kit- based GChemPaint application is being developed, as part of Bioclipse. Jmol Jmol applet is used to explore the Jmol is a free, open source molecule viewer structure of molecules. Jmol applet is for students, educators, and researchers used to depict X-ray structures in chemistry and biochemistry. It is cross- platform, running on Windows, Mac OS X, and Linux/Unix systems. For PG Students LaTeX Document markup language and Value addition to academic Skills set. preparation system for Tex typesetting Essential for International paper presentation and scientific journals. For PG student for their project work Scilab Scientific Computation package for Value addition in technical problem numerical computations solving via use of computational methods for engineering problems, Applicable in Chemical, ECE, Electrical, Electronics, Civil, Mechanical, Mathematics etc. For PG student who are taking Physical Chemistry Avogadro Avogadro is a free and open source, Research and Development in Chemistry, advanced molecule editor and Pharmacist and University lecturers. visualizer designed for cross-platform use in computational chemistry, molecular modeling, material science, bioinformatics, etc. Spoken Tutorial Project, IIT Bombay Brochure for Commerce and Commerce IT Name of FOSS Applications / Employability LibreOffice – Writer, Calc, Writing letters, documents, creating spreadsheets, tables, Impress making presentations, desktop publishing LibreOffice – Base, Draw, Managing databases, Drawing, doing simple Mathematical Math operations For Commerce IT Students Drupal Drupal is a free and open source content management system (CMS). -

Parameterizing a Novel Residue
University of Illinois at Urbana-Champaign Luthey-Schulten Group, Department of Chemistry Theoretical and Computational Biophysics Group Computational Biophysics Workshop Parameterizing a Novel Residue Rommie Amaro Brijeet Dhaliwal Zaida Luthey-Schulten Current Editors: Christopher Mayne Po-Chao Wen February 2012 CONTENTS 2 Contents 1 Biological Background and Chemical Mechanism 4 2 HisH System Setup 7 3 Testing out your new residue 9 4 The CHARMM Force Field 12 5 Developing Topology and Parameter Files 13 5.1 An Introduction to a CHARMM Topology File . 13 5.2 An Introduction to a CHARMM Parameter File . 16 5.3 Assigning Initial Values for Unknown Parameters . 18 5.4 A Closer Look at Dihedral Parameters . 18 6 Parameter generation using SPARTAN (Optional) 20 7 Minimization with new parameters 32 CONTENTS 3 Introduction Molecular dynamics (MD) simulations are a powerful scientific tool used to study a wide variety of systems in atomic detail. From a standard protein simulation, to the use of steered molecular dynamics (SMD), to modelling DNA-protein interactions, there are many useful applications. With the advent of massively parallel simulation programs such as NAMD2, the limits of computational anal- ysis are being pushed even further. Inevitably there comes a time in any molecular modelling scientist’s career when the need to simulate an entirely new molecule or ligand arises. The tech- nique of determining new force field parameters to describe these novel system components therefore becomes an invaluable skill. Determining the correct sys- tem parameters to use in conjunction with the chosen force field is only one important aspect of the process. -
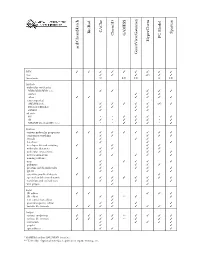
D:\Doc\Workshops\2005 Molecular Modeling\Notebook Pages\Software Comparison\Summary.Wpd
CAChe BioRad Spartan GAMESS Chem3D PC Model HyperChem acd/ChemSketch GaussView/Gaussian WIN TTTT T T T T T mac T T T (T) T T linux/unix U LU LU L LU Methods molecular mechanics MM2/MM3/MM+/etc. T T T T T Amber T T T other TT T T T T semi-empirical AM1/PM3/etc. T T T T T (T) T Extended Hückel T T T T ZINDO T T T ab initio HF * * T T T * T dft T * T T T * T MP2/MP4/G1/G2/CBS-?/etc. * * T T T * T Features various molecular properties T T T T T T T T T conformer searching T T T T T crystals T T T data base T T T developer kit and scripting T T T T molecular dynamics T T T T molecular interactions T T T T movies/animations T T T T T naming software T nmr T T T T T polymers T T T T proteins and biomolecules T T T T T QSAR T T T T scientific graphical objects T T spectral and thermodynamic T T T T T T T T transition and excited state T T T T T web plugin T T Input 2D editor T T T T T 3D editor T T ** T T text conversion editor T protein/sequence editor T T T T various file formats T T T T T T T T Output various renderings T T T T ** T T T T various file formats T T T T ** T T T animation T T T T T graphs T T spreadsheet T T T * GAMESS and/or GAUSSIAN interface ** Text only. -

Pymol Modelling Workshop
PyMOL Modelling Workshop My website: http://pldserver1.biochem.queensu.ca/~rlc/work/teaching/BCHM442/ There you will find links to download the latest educational version of PyMOL as well as a link to my “Introduction to PyMOL”, which in turn has links to other people's PyMOL tutorials. Note also the PyMOL Wiki: http://pymolwiki.org. Structure files can be found by searching the Protein Data Bank (PDB) for structure: easy to remember website http://www.pdb.org. There is also the PDBe (PDB Europe, http://www.pdbe.org) that contains the same databank of structures, but with a different web interface for searching for structures and a different set of tools for analyzing structures. What is in a PDB file? Lots of information in the “header” (the section of the file preceding the actual atomic coordinates) as well as the coordinates for the atoms. When assessing a structure, one needs to take account of the resolution and R-factor, error estimates and missing residues. There is information about the sequence that was used to determine the structure with a sequence database reference. There is also information about the biological unit. In the case of crystal structures the biological unit may need to be generated by applying crystallographic symmetry operators, although there are web sites that also try to provide that information. (e.g. http://www.ebi.ac.uk/msd-srv/pisa/). Warning: One cannot blindly trust a crystal structure to be providing you with a completely accurate picture of reality. Reading the paper that describes the structure is a good start! Outline of PyMOL usage PyMOL is a very powerful, scriptable (customizable) tool for making publication-quality figures and performing analyses on structures. -

Dmol Guide to Select a Dmol3 Task 1
DMOL3 GUIDE MATERIALS STUDIO 8.0 Copyright Notice ©2014 Dassault Systèmes. All rights reserved. 3DEXPERIENCE, the Compass icon and the 3DS logo, CATIA, SOLIDWORKS, ENOVIA, DELMIA, SIMULIA, GEOVIA, EXALEAD, 3D VIA, BIOVIA and NETVIBES are commercial trademarks or registered trademarks of Dassault Systèmes or its subsidiaries in the U.S. and/or other countries. All other trademarks are owned by their respective owners. Use of any Dassault Systèmes or its subsidiaries trademarks is subject to their express written approval. Acknowledgments and References To print photographs or files of computational results (figures and/or data) obtained using BIOVIA software, acknowledge the source in an appropriate format. For example: "Computational results obtained using software programs from Dassault Systèmes Biovia Corp.. The ab initio calculations were performed with the DMol3 program, and graphical displays generated with Materials Studio." BIOVIA may grant permission to republish or reprint its copyrighted materials. Requests should be submitted to BIOVIA Support, either through electronic mail to [email protected], or in writing to: BIOVIA Support 5005 Wateridge Vista Drive, San Diego, CA 92121 USA Contents DMol3 1 Setting up a molecular dynamics calculation20 Introduction 1 Choosing an ensemble 21 Further Information 1 Defining the time step 21 Tasks in DMol3 2 Defining the thermostat control 21 Energy 3 Constraints during dynamics 21 Setting up the calculation 3 Setting up a transition state calculation 22 Dynamics 4 Which method to use? -

Atomistic Molecular Dynamics Simulation of Graphene- Isoprene Nanocomposites
Atomistic molecular dynamics simulation of graphene- isoprene nanocomposites P Chanlert1, J Wong-Ekkabut2, W Liewrian3,4,5 and T Sutthibutpong3,4,5* 1 Program of Physics, Faculty of Science and Technology, Songkhla Rajabhat University, Songkhla, 90000, Thailand 2 Department of Physics, Faculty of Science, Kasetsart University, Bangkok 10900, Thailand 3 Theoretical and Computational Physics Group, Department of Physics, Faculty of Science, King Mongkut’s University of Technology Thonburi (KMUTT), Bangkok 10140, Thailand 4 Theoretical and Computational Science Center (TaCS), Science Laboratory Building, Faculty of Science, King Mongkut’s University of Technology Thonburi (KMUTT), Bangkok 10140, Thailand 5 Thailand Center of Excellence in Physics (ThEP Center), Commission on Higher Education, Bangkok 10400, Thailand *email: [email protected] Abstract. A series of atomistic molecular dynamics simulations were performed for 4-mer isoprene molecules confined between graphene sheets with varied graphene separations in order to observe the finite size effects arose from the van der Waals interactions between the isoprene oligomer, forming high density shells at the graphene interfaces. For the small confinement regions, 1.24 nm and 2.21 nm, local density of isoprene at the graphene interface becomes higher than 10 times the bulk density. These results provided the further insights towards the rational design of nanostructures based on graphene sheets and natural rubber polymer. 1. Introduction Composite material is the combination between two materials or more where chemical reaction between these materials is absent. In composites, one with higher quantity is called matrix while those with lesser quantity are called fillers. In general, adding fillers into matrix material enhances both physical and mechanical properties in some ways. -

Jaguar 5.5 User Manual Copyright © 2003 Schrödinger, L.L.C
Jaguar 5.5 User Manual Copyright © 2003 Schrödinger, L.L.C. All rights reserved. Schrödinger, FirstDiscovery, Glide, Impact, Jaguar, Liaison, LigPrep, Maestro, Prime, QSite, and QikProp are trademarks of Schrödinger, L.L.C. MacroModel is a registered trademark of Schrödinger, L.L.C. To the maximum extent permitted by applicable law, this publication is provided “as is” without warranty of any kind. This publication may contain trademarks of other companies. October 2003 Contents Chapter 1: Introduction.......................................................................................1 1.1 Conventions Used in This Manual.......................................................................2 1.2 Citing Jaguar in Publications ...............................................................................3 Chapter 2: The Maestro Graphical User Interface...........................................5 2.1 Starting Maestro...................................................................................................5 2.2 The Maestro Main Window .................................................................................7 2.3 Maestro Projects ..................................................................................................7 2.4 Building a Structure.............................................................................................9 2.5 Atom Selection ..................................................................................................10 2.6 Toolbar Controls ................................................................................................11 -

Porting the DFT Code CASTEP to Gpgpus
Porting the DFT code CASTEP to GPGPUs Toni Collis [email protected] EPCC, University of Edinburgh CASTEP and GPGPUs Outline • Why are we interested in CASTEP and Density Functional Theory codes. • Brief introduction to CASTEP underlying computational problems. • The OpenACC implementation http://www.nu-fuse.com CASTEP: a DFT code • CASTEP is a commercial and academic software package • Capable of Density Functional Theory (DFT) and plane wave basis set calculations. • Calculates the structure and motions of materials by the use of electronic structure (atom positions are dictated by their electrons). • Modern CASTEP is a re-write of the original serial code, developed by Universities of York, Durham, St. Andrews, Cambridge and Rutherford Labs http://www.nu-fuse.com CASTEP: a DFT code • DFT/ab initio software packages are one of the largest users of HECToR (UK national supercomputing service, based at University of Edinburgh). • Codes such as CASTEP, VASP and CP2K. All involve solving a Hamiltonian to explain the electronic structure. • DFT codes are becoming more complex and with more functionality. http://www.nu-fuse.com HECToR • UK National HPC Service • Currently 30- cabinet Cray XE6 system – 90,112 cores • Each node has – 2×16-core AMD Opterons (2.3GHz Interlagos) – 32 GB memory • Peak of over 800 TF and 90 TB of memory http://www.nu-fuse.com HECToR usage statistics Phase 3 statistics (Nov 2011 - Apr 2013) Ab initio codes (VASP, CP2K, CASTEP, ONETEP, NWChem, Quantum Espresso, GAMESS-US, SIESTA, GAMESS-UK, MOLPRO) GS2NEMO ChemShell 2%2% SENGA2% 3% UM Others 4% 34% MITgcm 4% CASTEP 4% GROMACS 6% DL_POLY CP2K VASP 5% 8% 19% http://www.nu-fuse.com HECToR usage statistics Phase 3 statistics (Nov 2011 - Apr 2013) 35% of the Chemistry software on HECToR is using DFT methods. -
Massive-Parallel Implementation of the Resolution-Of-Identity Coupled
Article Cite This: J. Chem. Theory Comput. 2019, 15, 4721−4734 pubs.acs.org/JCTC Massive-Parallel Implementation of the Resolution-of-Identity Coupled-Cluster Approaches in the Numeric Atom-Centered Orbital Framework for Molecular Systems † § † † ‡ § § Tonghao Shen, , Zhenyu Zhu, Igor Ying Zhang,*, , , and Matthias Scheffler † Department of Chemistry, Fudan University, Shanghai 200433, China ‡ Shanghai Key Laboratory of Molecular Catalysis and Innovative Materials, MOE Key Laboratory of Computational Physical Science, Fudan University, Shanghai 200433, China § Fritz-Haber-Institut der Max-Planck-Gesellschaft, Faradayweg 4-6, 14195 Berlin, Germany *S Supporting Information ABSTRACT: We present a massive-parallel implementation of the resolution of identity (RI) coupled-cluster approach that includes single, double, and perturbatively triple excitations, namely, RI-CCSD(T), in the FHI-aims package for molecular systems. A domain-based distributed-memory algorithm in the MPI/OpenMP hybrid framework has been designed to effectively utilize the memory bandwidth and significantly minimize the interconnect communication, particularly for the tensor contraction in the evaluation of the particle−particle ladder term. Our implementation features a rigorous avoidance of the on- the-fly disk storage and excellent strong scaling of up to 10 000 and more cores. Taking a set of molecules with different sizes, we demonstrate that the parallel performance of our CCSD(T) code is competitive with the CC implementations in state-of- the-art high-performance-computing computational chemistry packages. We also demonstrate that the numerical error due to the use of RI approximation in our RI-CCSD(T) method is negligibly small. Together with the correlation-consistent numeric atom-centered orbital (NAO) basis sets, NAO-VCC-nZ, the method is applied to produce accurate theoretical reference data for 22 bio-oriented weak interactions (S22), 11 conformational energies of gaseous cysteine conformers (CYCONF), and 32 Downloaded via FRITZ HABER INST DER MPI on January 8, 2021 at 22:13:06 (UTC). -

Lawrence Berkeley National Laboratory Recent Work
Lawrence Berkeley National Laboratory Recent Work Title From NWChem to NWChemEx: Evolving with the Computational Chemistry Landscape. Permalink https://escholarship.org/uc/item/4sm897jh Journal Chemical reviews, 121(8) ISSN 0009-2665 Authors Kowalski, Karol Bair, Raymond Bauman, Nicholas P et al. Publication Date 2021-04-01 DOI 10.1021/acs.chemrev.0c00998 Peer reviewed eScholarship.org Powered by the California Digital Library University of California From NWChem to NWChemEx: Evolving with the computational chemistry landscape Karol Kowalski,y Raymond Bair,z Nicholas P. Bauman,y Jeffery S. Boschen,{ Eric J. Bylaska,y Jeff Daily,y Wibe A. de Jong,x Thom Dunning, Jr,y Niranjan Govind,y Robert J. Harrison,k Murat Keçeli,z Kristopher Keipert,? Sriram Krishnamoorthy,y Suraj Kumar,y Erdal Mutlu,y Bruce Palmer,y Ajay Panyala,y Bo Peng,y Ryan M. Richard,{ T. P. Straatsma,# Peter Sushko,y Edward F. Valeev,@ Marat Valiev,y Hubertus J. J. van Dam,4 Jonathan M. Waldrop,{ David B. Williams-Young,x Chao Yang,x Marcin Zalewski,y and Theresa L. Windus*,r yPacific Northwest National Laboratory, Richland, WA 99352 zArgonne National Laboratory, Lemont, IL 60439 {Ames Laboratory, Ames, IA 50011 xLawrence Berkeley National Laboratory, Berkeley, 94720 kInstitute for Advanced Computational Science, Stony Brook University, Stony Brook, NY 11794 ?NVIDIA Inc, previously Argonne National Laboratory, Lemont, IL 60439 #National Center for Computational Sciences, Oak Ridge National Laboratory, Oak Ridge, TN 37831-6373 @Department of Chemistry, Virginia Tech, Blacksburg, VA 24061 4Brookhaven National Laboratory, Upton, NY 11973 rDepartment of Chemistry, Iowa State University and Ames Laboratory, Ames, IA 50011 E-mail: [email protected] 1 Abstract Since the advent of the first computers, chemists have been at the forefront of using computers to understand and solve complex chemical problems. -

Computational Chemistry
Computational Chemistry as a Teaching Aid Melissa Boule Aaron Harshman Rexford Hoadley An Interactive Qualifying Project Report Submitted to the Faculty of the Worcester Polytechnic Institute In partial fulfillment of the requirements for the Bachelor of Science Degree Approved by: Professor N. Aaron Deskins 1i Abstract Molecular modeling software has transformed the capabilities of researchers. Molecular modeling software has the potential to be a helpful teaching tool, though as of yet its efficacy as a teaching technique has yet to be proven. This project focused on determining the effectiveness of a particular molecular modeling software in high school classrooms. Our team researched what topics students struggled with, surveyed current high school chemistry teachers, chose a modeling software, and then developed a lesson plan around these topics. Lastly, we implemented the lesson in two separate high school classrooms. We concluded that these programs show promise for the future, but with the current limitations of high school technology, these tools may not be as impactful. 2ii Acknowledgements This project would not have been possible without the help of many great educators. Our advisor Professor N. Aaron Deskins provided driving force to ensure the research ran smoothly and efficiently. Mrs. Laurie Hanlan and Professor Drew Brodeur provided insight and guidance early in the development of our topic. Mr. Mark Taylor set-up our WebMO server and all our other technology needs. Lastly, Mrs. Pamela Graves and Mr. Eric Van Inwegen were instrumental by allowing us time in their classrooms to implement our lesson plans. Without the support of this diverse group of people this project would never have been possible.