An Ethoxyquin-Inducible Aldehyde Reductase from Rat Liver That
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Datasheet: VMA00346 Product Details
Datasheet: VMA00346 Description: MOUSE ANTI AKR1C2 Specificity: AKR1C2 Format: Purified Product Type: PrecisionAb™ Monoclonal Isotype: IgG2a Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Product Form Purified IgG - liquid Preparation Mouse monoclonal antibody prepared by affinity chromatography on Protein G Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers Immunogen Recombinant human AKR1C2 External Database Links UniProt: P52895 Related reagents Entrez Gene: 1646 AKR1C2 Related reagents Synonyms DDH2 Specificity Mouse anti Human AKR1C2 antibody recognizes the aldo-keto reductase family 1 member C2, also known as 3-alpha-HSD3, DD-2, DD/BABP, aldo-keto reductase family 1 member C2, Page 1 of 2 chlordecone reductase homolog HAKRD, dihydrodiol dehydrogenase 2, bile acid binding protein, 3-alpha hydroxysteroid dehydrogenase, type III, pseudo-chlordecone reductase, testicular 17,20-desmolase deficiency, trans-1,2-dihydrobenzene-1,2-diol dehydrogenase and type II dihydrodiol dehydrogenase. Encoded by the AKR1C2 gene, aldo-keto reductase family 1 member C2 is a member of the aldo/keto reductase superfamily, which consists of more than 40 known enzymes and proteins. -

Datasheet: VMA00334 Product Details
Datasheet: VMA00334 Description: MOUSE ANTI AKR1C1 Specificity: AKR1C1 Format: Purified Product Type: PrecisionAb™ Monoclonal Isotype: IgG1 Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Product Form Purified IgG - liquid Preparation Mouse monoclonal antibody prepared by affinity chromatography on Protein G Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers Immunogen Recombinant human AKR1C1 External Database Links UniProt: Q04828 Related reagents Entrez Gene: 1645 AKR1C1 Related reagents Synonyms DDH, DDH1 Specificity Mouse anti Human AKR1C1 antibody detects the aldo-keto reductase family 1 member C1, also known as 20-alpha-hydroxysteroid dehydrogenase (20-alpha-HSD), chlordecone reductase Page 1 of 2 homolog HAKRC and high-affinity hepatic bile acid-binding protein (HBAB). Encoded by the AKR1C1 gene, aldo-keto reductase family 1 member C1 is a member of the aldo/keto reductase superfamily, which consists of more than 40 known enzymes and proteins. These enzymes catalyze the conversion of aldehydes and ketones to their corresponding alcohols by utilizing NADH and/or NADPH as cofactors. -

Aldo-Keto Reductase 1C3 Recombinant Protein Cat
Aldo-Keto Reductase 1C3 Recombinant Protein Cat. No.: 91-710 Aldo-Keto Reductase 1C3 Recombinant Protein Specifications SPECIES: Human SOURCE SPECIES: Human Cells SEQUENCE: Met1-Tyr323 FUSION TAG: C-6 His tag TESTED APPLICATIONS: APPLICATIONS: This recombinant protein can be used for biological assays. For research use only. PREDICTED MOLECULAR 37.8 kD WEIGHT: Properties Greater than 95% as determined by reducing SDS-PAGE. PURITY: Endotoxin level less than 0.1 ng/ug (1 IEU/ug) as determined by LAL test. PHYSICAL STATE: Lyophilized Lyophilized from a 0.2 um filtered solution of 20mM PB,150mM NaCl,pH7.4. It is not BUFFER: recommended to reconstitute to a concentration less than 100 ug/ml. Dissolve the lyophilized protein in ddH2O. September 23, 2021 1 https://www.prosci-inc.com/aldo-keto-reductase-1c3-recombinant-protein-91-710.html Lyophilized protein should be stored at -20˚C, though stable at room temperature for 3 weeks. STORAGE CONDITIONS: Reconstituted protein solution can be stored at 4-7˚C for 2-7 days. Aliquots of reconstituted samples are stable at -20˚C for 3 months. Additional Info OFFICIAL SYMBOL: AKR1C3 Aldo-Keto Reductase Family 1 Member C3, 17-Beta-Hydroxysteroid Dehydrogenase Type 5, 17-Beta-HSD 5, 3-Alpha-HSD Type II Brain, 3-Alpha-Hydroxysteroid Dehydrogenase Type 2, 3-Alpha-HSD Type 2, Chlordecone Reductase Homolog HAKRb, Dihydrodiol ALTERNATE NAMES: Dehydrogenase 3, DD-3, DD3, Dihydrodiol Dehydrogenase Type I, HA1753, Indanol Dehydrogenase, Prostaglandin F Synthase, Testosterone 17-Beta-Dehydrogenase 5, Trans-1, 2-Dihydrobenzene-1, 2-Diol Dehydrogenase, AKR1C3, DDH1, HSD17B5, KIAA0119, PGFS ACCESSION NO.: P42330 GENE ID: 8644 Background and References AKR1C3, is an enzyme which belongs to the aldo/keto reductase family. -

Table 3: Average Gene Expression Profiles by Chromosome
Supplemental Data Table 1: Experimental Setup Correlation Array Reverse Fluor Array Extraction Coefficient Print Batch (Y/N) mean (range) DLD1-I.1 I A N DLD1-I.2 I B N 0.86 DLD1-I.3 I C N (0.79-0.90) DLD1-I.4 I C Y DLD1 DLD1-II.1 II D N DLD1-II.2 II E N 0.86 DLD1-II.3 II F N (0.74-0.94) DLD1-II.4 II F Y DLD1+3-II.1 II A N DLD1+3-II.2 II A N 0.85 DLD1 + 3 DLD1+3-II.3 II B N (0.64-0.95) DLD1+3-II.4 II B Y DLD1+7-I.1 I A N DLD1+7-I.2 I A N 0.79 DLD1 + 7 DLD1+7-I.3 I B N (0.68-0.90) DLD1+7-I.4 I B Y DLD1+13-I.1 I A N DLD1+13-I.2 I A N 0.88 DLD1 + 13 DLD1+13-I.3 I B N (0.84-0.91) DLD1+13-I.4 I B Y hTERT-HME-I.1 I A N hTERT-HME-I.2 I B N 0.85 hTERT-HME hTERT-HME-I.3 I C N (0.80-0.92) hTERT-HME-I.4 I C Y hTERT-HME+3-I.1 I A N hTERT-HME+3-I.2 I B N 0.84 hTERT-HME + 3 hTERT-HME+3-I.3 I C N (0.74-0.90) hTERT-HME+3-I.4 I C Y Supplemental Data Table 2: Average gene expression profiles by chromosome arm DLD1 hTERT-HME Ratio.7 Ratio.1 Ratio.3 Ratio.3 Chrom. -

Protein T1 C1 Accession No. Description
Protein T1 C1 Accession No. Description SW:143B_HUMAN + + P31946 14-3-3 protein beta/alpha (protein kinase c inhibitor protein-1) (kcip-1) (protein 1054). 14-3-3 protein epsilon (mitochondrial import stimulation factor l subunit) (protein SW:143E_HUMAN + + P42655 P29360 Q63631 kinase c inhibitor protein-1) (kcip-1) (14-3-3e). SW:143S_HUMAN + - P31947 14-3-3 protein sigma (stratifin) (epithelial cell marker protein 1). SW:143T_HUMAN + - P27348 14-3-3 protein tau (14-3-3 protein theta) (14-3-3 protein t-cell) (hs1 protein). 14-3-3 protein zeta/delta (protein kinase c inhibitor protein-1) (kcip-1) (factor SW:143Z_HUMAN + + P29312 P29213 activating exoenzyme s) (fas). P01889 Q29638 Q29681 Q29854 Q29861 Q31613 hla class i histocompatibility antigen, b-7 alpha chain precursor (mhc class i antigen SW:1B07_HUMAN + - Q9GIX1 Q9TP95 b*7). hla class i histocompatibility antigen, b-14 alpha chain precursor (mhc class i antigen SW:1B14_HUMAN + - P30462 O02862 P30463 b*14). P30479 O19595 Q29848 hla class i histocompatibility antigen, b-41 alpha chain precursor (mhc class i antigen SW:1B41_HUMAN + - Q9MY79 Q9MY94 b*41) (bw-41). hla class i histocompatibility antigen, b-42 alpha chain precursor (mhc class i antigen SW:1B42_HUMAN + - P30480 P79555 b*42). P30488 O19615 O19624 O19641 O19783 O46702 hla class i histocompatibility antigen, b-50 alpha chain precursor (mhc class i antigen SW:1B50_HUMAN + - O78172 Q9TQG1 b*50) (bw-50) (b-21). hla class i histocompatibility antigen, b-54 alpha chain precursor (mhc class i antigen SW:1B54_HUMAN + - P30492 Q9TPQ9 b*54) (bw-54) (bw-22). P30495 O19758 P30496 hla class i histocompatibility antigen, b-56 alpha chain precursor (mhc class i antigen SW:1B56_HUMAN - + P79490 Q9GIM3 Q9GJ17 b*56) (bw-56) (bw-22). -

Supplemental Figures 04 12 2017
Jung et al. 1 SUPPLEMENTAL FIGURES 2 3 Supplemental Figure 1. Clinical relevance of natural product methyltransferases (NPMTs) in brain disorders. (A) 4 Table summarizing characteristics of 11 NPMTs using data derived from the TCGA GBM and Rembrandt datasets for 5 relative expression levels and survival. In addition, published studies of the 11 NPMTs are summarized. (B) The 1 Jung et al. 6 expression levels of 10 NPMTs in glioblastoma versus non‐tumor brain are displayed in a heatmap, ranked by 7 significance and expression levels. *, p<0.05; **, p<0.01; ***, p<0.001. 8 2 Jung et al. 9 10 Supplemental Figure 2. Anatomical distribution of methyltransferase and metabolic signatures within 11 glioblastomas. The Ivy GAP dataset was downloaded and interrogated by histological structure for NNMT, NAMPT, 12 DNMT mRNA expression and selected gene expression signatures. The results are displayed on a heatmap. The 13 sample size of each histological region as indicated on the figure. 14 3 Jung et al. 15 16 Supplemental Figure 3. Altered expression of nicotinamide and nicotinate metabolism‐related enzymes in 17 glioblastoma. (A) Heatmap (fold change of expression) of whole 25 enzymes in the KEGG nicotinate and 18 nicotinamide metabolism gene set were analyzed in indicated glioblastoma expression datasets with Oncomine. 4 Jung et al. 19 Color bar intensity indicates percentile of fold change in glioblastoma relative to normal brain. (B) Nicotinamide and 20 nicotinate and methionine salvage pathways are displayed with the relative expression levels in glioblastoma 21 specimens in the TCGA GBM dataset indicated. 22 5 Jung et al. 23 24 Supplementary Figure 4. -
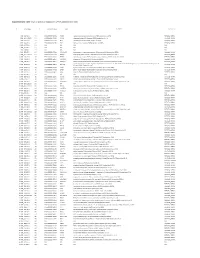
Supplementary Table 1. List of Genes Up-Regulated in LPAR6 Knocked Down Cells
Supplementary Table 1. List of genes up-regulated in LPAR6 knocked down cells g#initial alias c# converted alias name description namespace 1 NM_004132.3 1.1 ENSG00000148702 HABP2 hyaluronan binding protein 2 [Source:HGNC Symbol;Acc:4798] REFSEQ_MRNA 2 NM_001142282.1 2.1 ENSG00000138326 RPS24 ribosomal protein S24 [Source:HGNC Symbol;Acc:10411] REFSEQ_MRNA 3 NM_000301.3 3.1 ENSG00000122194 PLG plasminogen [Source:HGNC Symbol;Acc:9071] REFSEQ_MRNA 4 NM_004467.3 4.1 ENSG00000104760 FGL1 fibrinogen-like 1 [Source:HGNC Symbol;Acc:3695] REFSEQ_MRNA 5 NR_015379.3 5.1 N/A N/A N/A N/A 6 NM_001958.3 6.1 N/A N/A N/A N/A 7 NG_008956.1 7.1 N/A N/A N/A N/A 8 NM_033520.1 8.1 ENSG00000167644 C19ORF33 chromosome 19 open reading frame 33 [Source:HGNC Symbol;Acc:16668] REFSEQ_MRNA 9 NM_001242946.1 9.1 ENSG00000086289 EPDR1 ependymin related protein 1 (zebrafish) [Source:HGNC Symbol;Acc:17572] REFSEQ_MRNA 10 NM_176796.1 10.1 ENSG00000171631 P2RY6 pyrimidinergic receptor P2Y, G-protein coupled, 6 [Source:HGNC Symbol;Acc:8543] REFSEQ_MRNA 11 NM_178562.3 11.1 ENSG00000158457 TSPAN33 tetraspanin 33 [Source:HGNC Symbol;Acc:28743] REFSEQ_MRNA 12 NM_198488.3 12.1 ENSG00000180921 FAM83H family with sequence similarity 83, member H [Source:HGNC Symbol;Acc:24797] REFSEQ_MRNA aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4) 13 NM_001818.3 13.1 ENSG00000198610 AKR1C4 REFSEQ_MRNA [Source:HGNC Symbol;Acc:387] 14 NM_001311.4 14.1 ENSG00000213145 CRIP1 cysteine-rich protein 1 (intestinal) -

Monoclonal Antibody to AKR1C1 / DHH1 (1-323) - Purified
OriGene Technologies, Inc. OriGene Technologies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] AM09389PU-S Monoclonal Antibody to AKR1C1 / DHH1 (1-323) - Purified Alternate names: 2-dihydrobenzene-1, 2-diol dehydrogenase, 20-alpha-hydroxysteroid dehydrogenase, Aldo-keto reductase family 1 member C1, Chlordecone reductase homolog HAKRC, Dihydrodiol dehydrogenase 1/2, High-affinity hepatic bile acid-binding protein, Trans-1 Quantity: 50 µl Concentration: 1.0 mg/ml Background: The human aldo-keto reductases 1C1 and 1C3 (AKR1C1 and AKR1C3) have major roles in pre receptor regulation of progesterone action. They can both convert progesterone to the less potent efficiencies. AKR1C1 and AKR1C3 also act as 3-ketosteroid reductase, and as such they can convert the most potent androgen 5alpha-DHT into 3beta- andorstandiol, which is an estrogen receptor beta ligand, and into the inactive androgen 3alpha-androstnionl, respectively. Uniprot ID: Q04828 NCBI: NP_001344 GeneID: 1645 Host / Isotype: Mouse / IgG1 Recommended Isotype SM10P (for use in human samples), AM03095PU-N Controls: Clone: AT6D10 Immunogen: Recombinant Human AKR1C1 (1-323aa) purified from E. coli Format: State: Liquid purified Ig fraction Purification: Affinity Chromatography on Protein G Buffer System: PBS, pH 7.4 containing 0.02% Sodium Azide and 10% Glycerol Applications: ELISA. Western blot: 1/1000. Immunohistochemistry on Paraffin Sections: 10 µg/ml. Immunocytochemistry / Immunoflourescence. Flow cytometry. Other applications not tested. Optimal dilutions are dependent on conditions and should be determined by the user. -

AKR1C3) in Neuroendocrine Tumors & Adenocarcinomas of Pancreas, Gastrointestinal Tract, and Lung
Int J Clin Exp Pathol 2013;6(11):2419-2429 www.ijcep.com /ISSN:1936-2625/IJCEP1309055 Original Article Expression of aldo-keto reductase family 1 member C3 (AKR1C3) in neuroendocrine tumors & adenocarcinomas of pancreas, gastrointestinal tract, and lung Theodore S Chang1, Hsueh-Kung Lin2, Kyle A Rogers3, Lacy S Brame4, Matthew M Yeh5, Qing Yang2, Kar-Ming Fung1,2,4,6 Departments of 1Pathology, 2Urology, 3College of Medicine, 4Stephenson Cancer Center, University of Oklahoma Health Sciences Center, Oklahoma City, OK; 5Department of Pathology, University of Washington School of Medi- cine, Seattle, WA; 6Department of Pathology, Veterans Administration Medical Center, Oklahoma City, OK Received September 22, 2013; Accepted October 15, 2013; Epub October 15, 2013; Published November 1, 2013 Abstract: Human aldo-keto reductase family 1 member C3 (AKR1C3) was initially identified as an enzyme in reduc- ing 5α-dihydrotestosterone (5α-DHT) to 5α-androstane-3α, 17β-diol (3α-diol) and oxidizing 3α-diol to androsterone. It was subsequently demonstrated to possess ketosteroid reductase activity in metabolizing other steroids including estrogen and progesterone, 11-ketoprostaglandin reductase activity in metabolizing prostaglandins, and dihydrodiol dehydrogenase x (DDx) activity in metabolizing xenobiotics. AKR1C3 was demonstrated in sex hormone-dependent tissues including testis, breast, endometrium, and prostate; in sex hormone-independent tissues including kidney and urothelium. Our previous study described the expression of AKR1C3 in squamous cell carcinoma and ad- enocarcinoma but not in small cell carcinoma. In this report, we studied the expression of AKR1C3 in normal tis- sue, adenocarcinomas (43 cases) and neuroendocrine (NE) tumors (40 cases) arising from the aerodigestive tract and pancreas. -

Human Aldo-Keto Reductase 1C1/AKR1C1 Antibody Catalog Number: ATGA0201
Human Aldo-keto Reductase 1C1/AKR1C1 antibody Catalog Number: ATGA0201 PRODUCT INPORMATION Catalog number ATGA0201 Clone No. AT6D10 Product type Monoclonal Antibody UnitProt No. Q04828 NCBI Accession No. NP_001344 Alternative Names Aldo-keto reductase family 1 member C1, 20-alpha-HSD, DD1/DD2, HBAB, DDH, DDH1, Aldo-keto reductase family 1, member C1 20 alpha (3 alpha) hydroxysteroid dehydrogenase, 20 alpha hydroxysteroid dehydrogenase, 2ALPHAHSD, AK1C1, MBAB, Aldo keto reductase family 1 member C1, C9, Chlordecone reductase homolog, Chlordecone reductase homolog HAKRC, DD1, Dihydrodiol dehydrogenase 1, Dihydrodiol dehydrogenase 1/2, H37, HAKRC, Hepatic dihydrodiol dehydrogenase, High affinity hepatic bile acid-binding protein, Trans-1,2 dihydrobenzene 1,2 diol dehydrogenase, Type II 3 alpha hydroxysteroid dehydrogenase, PRODUCT SPECIFICATION Antibody Host Mouse Reacts With Human Concentration 1mg/ml (determined by BCA assay) Formulation Liquid in. Phosphate-Buffered Saline (pH 7.4) with 0.02% Sodium Azide, 10% glycerol Immunogen Recombinant human AKR1C1 (1-323aa) purified from E. coli Isotype IgG1 kappa Purification Note By protein-G affinity chromatography Application ELISA, WB, ICC/IF, FACS 1 Human Aldo-keto Reductase 1C1/AKR1C1 antibody Catalog Number: ATGA0201 Usage The antibody has been tested by ELISA, Western blot, ICC/IF and FACS analysis to assure specificity and reactivity. Since application varies, however, each investigation should be titrated by the reagent to obtain optimal results. Storage Can be stored at +2C to +8C for 1 week. For long term storage, aliquot and store at -20C to -80C. Avoid repeated freezing and thawing cycles. BACKGROUND Description The human aldo-keto reductases 1C1 and 1C3 (AKR1C1 and AKR1C3) have major roles in pre receptor regulation of progesterone action. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

Selective AKR1C3 Inhibitors Do Not Recapitulate the Anti-Leukaemic Activities of the Pan-AKR1C Inhibitor Medroxyprogesterone Acetate
FULL PAPER British Journal of Cancer (2014) 110, 1506–1516 | doi: 10.1038/bjc.2014.83 Keywords: aldo-keto reductase 1C3; inhibitors; leukaemia Selective AKR1C3 inhibitors do not recapitulate the anti-leukaemic activities of the pan-AKR1C inhibitor medroxyprogesterone acetate F Khanim1,8, N Davies2,8, P Velic¸a3, R Hayden1, J Ride1, C Pararasa4, M G Chong2, U Gunther2, N Veerapen1, P Winn5, R Farmer5, E Trivier6, L Rigoreau6, M Drayson7 and C Bunce*,1 1School of Biosciences, University of Birmingham, Birmingham B15 2TT, UK; 2School of Cancer Sciences, University of Birmingham, Birmingham B15 2TT, UK; 3Haematology Department, UCL Cancer Institute, London WC1E 6DD, UK; 4School of Life and Health Sciences, Aston University, Birmingham B4 7ET, UK; 5Centre for Systems Biology, University of Birmingham, Birmingham B15 2TT, UK; 6CRT Discovery Laboratories, Babraham, Cambridge CB22 3AT, UK and 7Immunity and Infection, University of Birmingham, Birmingham B15 2TT, UK Background: We and others have identified the aldo-keto reductase AKR1C3 as a potential drug target in prostate cancer, breast cancer and leukaemia. As a consequence, significant effort is being invested in the development of AKR1C3-selective inhibitors. Methods: We report the screening of an in-house drug library to identify known drugs that selectively inhibit AKR1C3 over the closely related isoforms AKR1C1, 1C2 and 1C4. This screen initially identified tetracycline as a potential AKR1C3-selective inhibitor. However, mass spectrometry and nuclear magnetic resonance studies identified that the active agent was a novel breakdown product (4-methyl(de-dimethylamine)-tetracycline (4-MDDT)). Results: We demonstrate that, although 4-MDDT enters AML cells and inhibits their AKR1C3 activity, it does not recapitulate the anti-leukaemic actions of the pan-AKR1C inhibitor medroxyprogesterone acetate (MPA).