Prof. Yi-Xue Li
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Cantonese for Helpers
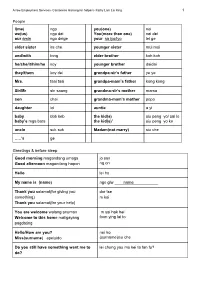
Arrow Employment Services- Cantonese learning for helpers- Kathy Lam Lai King 1 People I(me) ngo you(one) nei we(us) ngo dei You(more than one) nei dei our amin ngo deige your sa iyo/iyo lei ge elder sister ka che younger sister mui mui and/with tong elder brother koh koh he/she/it/him/he koy younger brother daidai they/them koy dei grandpa-sir’s father ye ye Mrs. taai taai grandpa-mam’s father kong kong Sir/Mr sin saang grandma-sir’s mother mama son chai grandma-mam’s mother popo daughter loi auntie a yi baby bbb keb the kid(s) siu peng yo/ sai lo baby’s mga bata the kid(s)’ siu peng yo ke uncle suk suk Madam(not marry) siu che …..’s ge Greetings & before sleep Good morning magandang umaga jo san Good afternoon magandang hapon ng on Hello lei ho My name is (name) ngo giw ____name___________ Thank you salamat(for giving you dor tse something) m koi Thank you salamat(for your help) You are welcome walang anuman m sai hak hei Welcome to this home maligayang foon ying lai to pagdating Hello/How are you? nei ho Miss(surname) apelyido (surname)siu che Do you still have something want me to lei chung yau mo kei ta fan fu? do? Arrow Employment Services- Cantonese learning for helpers- Kathy Lam Lai King 2 If not, I will take shower yue kuo mo, ngo heui chung leung Good night man ngong Common Questions/Answers Why bakit Where saan When kailan How paano what? tim kai hai bin to kei si tim yeng mak ye? How many/how much How long(time) How long(length) What time… yes ilan/magkano gaano katagal? gaano kahaba? anog oras? haih/yau kei to kei loi kei cheng kei to dim? What is it? ano yan Can I do it later? may be no/not li di hai mak ye? ngo hoh m hoh yi. -

1 Yi Guo, Pharmd, NYCSHP, President
President’s Message ………………………………………………………………......……...1 Yi Guo, PharmD, NYCSHP, President President-Elect’s Message…………………………………………………………....……......2 Jason Babby, PharmD, NYCSHP, President-Elect Current and Emerging Treatment for Pediatric Mitochondrial Disorders... …....…..................3 Kolaleh Hassan, MS, PharmD Candidate 2016 Autism and Gastrointestinal Disorders: Is there a link?...…………………..………......…......5 Kirolous Makarious, PharmD Candidate 2018 Herbal Products and the Pregnant Patient - Do Benefits Outweigh the Risks?..........................7 Michele Kaufman, PharmD, CGP Expedited Partner Therapy (EPT): A Strategic Approach for Preventative Therapy.......……9 Fatema Elias, PharmD Candidate 2016 Jeffrey Legaspi, PharmD Candidate 2016 Photo Galleries…………………………………………………………………......…....…...10 Volume XXXX Number 6 office at the time, it was inevitable that we darbepoetin injection and ribavirin for an discussed our lives at work and beyond. anemic patient at a HIV/Hepatitis C Under his influence, I decided to attend Pharmacotherapy clinic during my PGY2 the Thursday night Board of Directors Infectious Diseases training at the Bronx meetings which subsequently led me to VA, and the moment when I became my very first Annual Assembly Conference credentialed as an infectious disease in Saratoga. It was an eye-opening clinical pharmacist with a physician experience for me! To be honest, I was a collaborative agreement at Montefiore little overwhelmed: there were heated Medical Center. discussions and debates on “resolutions, I believe I don’t need to emphasize the CDTMs, and bylaws”; networking with importance of collaborative drug therapy President’s Message: members from other chapters; and Yi Guo, PharmD industry exhibits. This experience truly management (CDTM) and pharmacist sparked my interest to get more involved provider status to you all because we are and learn more of what we can do all in agreement. -

Lǎoshī Hé Xuéshēng (Teacher and Students)
© Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. CHAPTER Lǎoshī hé Xuéshēng 1 (Teacher and Students) Pinyin Text English Translation (A—Dīng Yī, B—Wáng Èr, C—Zhāng Sān) (A—Ding Yi, B—Wang Er, C—Zhang San) A: Nínhǎo, nín guìxìng? A: Hello, what is your honorable surname? B: Wǒ xìng Wáng, jiào Wáng Èr. Wǒ shì B: My surname is Wang. I am called Wang Er. lǎoshī. Nǐ xìng shénme? I am a teacher. What is your last name? A: Wǒ xìng Dīng, wǒde míngzi jiào Dīng Yī. A: My last name is Ding and my full name is Wǒ shì xuéshēng. Ding Yi. I am a student. 老师 老師 lǎoshī n. teacher 和 hé conj. and 学生 學生 xuéshēng n. student 您 nín pron. honorific form of singular you 好 hǎo adj. good 你(您)好 nǐ(nín)hǎo greeting hello 贵 貴 guì adj. honorable 贵姓 貴姓 guìxìng n./v. honorable surname (is) 我 wǒ pron. I; me 姓 xìng n./v. last name; have the last name of … 王 Wáng n. last name Wang 叫 jiào v. to be called 二 èr num. two (used when counting; here used as a name) 是 shì v. to be (any form of “to be”) 10 © Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. -

Ideophones in Middle Chinese
KU LEUVEN FACULTY OF ARTS BLIJDE INKOMSTSTRAAT 21 BOX 3301 3000 LEUVEN, BELGIË ! Ideophones in Middle Chinese: A Typological Study of a Tang Dynasty Poetic Corpus Thomas'Van'Hoey' ' Presented(in(fulfilment(of(the(requirements(for(the(degree(of(( Master(of(Arts(in(Linguistics( ( Supervisor:(prof.(dr.(Jean=Christophe(Verstraete((promotor)( ( ( Academic(year(2014=2015 149(431(characters Abstract (English) Ideophones in Middle Chinese: A Typological Study of a Tang Dynasty Poetic Corpus Thomas Van Hoey This M.A. thesis investigates ideophones in Tang dynasty (618-907 AD) Middle Chinese (Sinitic, Sino- Tibetan) from a typological perspective. Ideophones are defined as a set of words that are phonologically and morphologically marked and depict some form of sensory image (Dingemanse 2011b). Middle Chinese has a large body of ideophones, whose domains range from the depiction of sound, movement, visual and other external senses to the depiction of internal senses (cf. Dingemanse 2012a). There is some work on modern variants of Sinitic languages (cf. Mok 2001; Bodomo 2006; de Sousa 2008; de Sousa 2011; Meng 2012; Wu 2014), but so far, there is no encompassing study of ideophones of a stage in the historical development of Sinitic languages. The purpose of this study is to develop a descriptive model for ideophones in Middle Chinese, which is compatible with what we know about them cross-linguistically. The main research question of this study is “what are the phonological, morphological, semantic and syntactic features of ideophones in Middle Chinese?” This question is studied in terms of three parameters, viz. the parameters of form, of meaning and of use. -

IUSSP Laureate Nomination Letter for ZENG Yi
IUSSP Laureate Nomination letter for ZENG Yi We are pleased to nominate Professor Yi Zeng for his outstanding contributions to the advancement of population sciences and distinguished services rendered to the community of scholars in population sciences in general and IUSSP in particular. Yi Zeng’s contributions to the advancement of population science may be grouped into two categories: (a) research and (b) capacity building and institution building. In addition, we highlight service rendered to the IUSSP (section c). Yi Zeng’s contributions and service to the IUSSP and the scientific community at large have been acknowledged widely. A brief account of the recognition is offered in section d. a. Research During his professional career, Yi Zeng developed a research focus on healthy aging and family and household demography. In important ways, he shaped two areas that are central to population sciences. a.1 Healthy aging In 1998, he and colleagues in China, Europe and the United States launched the ground- breaking Chinese Longitudinal Healthy Longevity Study (CLHLS). The objective was to develop a knowledge base for enhancing the wellbeing and quality of life of elderly in China and worldwide. China was aging at an extraordinary pace and had the largest number of elderly persons in the world. The CLHLS was and is the largest longitudinal study of the oldest-old aged 80+ (with compatible young-old aged 65-79) in the world. Several innovative features were introduced to disentangle the effects of socioeconomic and behavioral/lifestyle factors, social support, environmental factors, genetics and genetics-environment interactions on healthy longevity. -

First Person – Yi Liu
© 2020. Published by The Company of Biologists Ltd | Journal of Cell Science (2020) 133, jcs246173. doi:10.1242/jcs.246173 FIRST PERSON First person – Yi Liu First Person is a series of interviews with the first authors of a selection of papers published in Journal of Cell Science, helping early-career researchers promote themselves alongside their papers. Yi Liu is first author on ‘Direct interaction between CEP85 and STIL mediates PLK4-driven directed cell migration’, published in JCS. Yi conducted the research described in this article while a PhD student in Dr Laurence Pelletier’s lab at Lunenfeld-Tanenbaum Research Institute, University of Toronto, Canada. He is now a Postdoc in the lab of Dr Rudolf Jaenisch at Whitehead Institute for Biomedical Research, MIT, USA, where he uses stem cells to study complex human brain disorders. How would you explain the main findings of your paper in lay terms? Metastasis is of paramount importance in the prognosis of cancer patients. Malignant cancer cells acquire the motile ability to invade the surrounding tissues and penetrate the vascular circulation to produce secondary tumors, which is the major cause of mortality of most cancer patients. The PLK4 gene has recently been proposed as a prime target for cancer therapeutics since its upregulation has a great impact on cancer metastasis. Hence, our work aims to characterize the molecular basis of PLK4-driven cancer cell motility. We found that CEP85 and STIL proteins form a complex that regulates PLK4 activity and thereby Yi Liu plays a critical role in cancer cell migration. Molecularly, this complex was shown to modulate the organization of actin, a major type of Why did you choose Journal of Cell Science for your paper? cytoskeleton that controls cell motility. -

HÀNWÉN and TAIWANESE SUBJECTIVITIES: a GENEALOGY of LANGUAGE POLICIES in TAIWAN, 1895-1945 by Hsuan-Yi Huang a DISSERTATION S
HÀNWÉN AND TAIWANESE SUBJECTIVITIES: A GENEALOGY OF LANGUAGE POLICIES IN TAIWAN, 1895-1945 By Hsuan-Yi Huang A DISSERTATION Submitted to Michigan State University in partial fulfillment of the requirements for the degree of Curriculum, Teaching, and Educational Policy—Doctor of Philosophy 2013 ABSTRACT HÀNWÉN AND TAIWANESE SUBJECTIVITIES: A GENEALOGY OF LANGUAGE POLICIES IN TAIWAN, 1895-1945 By Hsuan-Yi Huang This historical dissertation is a pedagogical project. In a critical and genealogical approach, inspired by Foucault’s genealogy and effective history and the new culture history of Sol Cohen and Hayden White, I hope pedagogically to raise awareness of the effect of history on shaping who we are and how we think about our self. I conceptualize such an historical approach as effective history as pedagogy, in which the purpose of history is to critically generate the pedagogical effects of history. This dissertation is a genealogical analysis of Taiwanese subjectivities under Japanese rule. Foucault’s theory of subjectivity, constituted by the four parts, substance of subjectivity, mode of subjectification, regimen of subjective practice, and telos of subjectification, served as a conceptual basis for my analysis of Taiwanese practices of the self-formation of a subject. Focusing on language policies in three historical events: the New Culture Movement in the 1920s, the Taiwanese Xiāngtǔ Literature Movement in the early 1930s, and the Japanization Movement during Wartime in 1937-1945, I analyzed discourses circulating within each event, particularly the possibilities/impossibilities created and shaped by discourses for Taiwanese subjectification practices. I illustrate discursive and subjectification practices that further shaped particular Taiwanese subjectivities in a particular event. -

Names of Chinese People in Singapore
101 Lodz Papers in Pragmatics 7.1 (2011): 101-133 DOI: 10.2478/v10016-011-0005-6 Lee Cher Leng Department of Chinese Studies, National University of Singapore ETHNOGRAPHY OF SINGAPORE CHINESE NAMES: RACE, RELIGION, AND REPRESENTATION Abstract Singapore Chinese is part of the Chinese Diaspora.This research shows how Singapore Chinese names reflect the Chinese naming tradition of surnames and generation names, as well as Straits Chinese influence. The names also reflect the beliefs and religion of Singapore Chinese. More significantly, a change of identity and representation is reflected in the names of earlier settlers and Singapore Chinese today. This paper aims to show the general naming traditions of Chinese in Singapore as well as a change in ideology and trends due to globalization. Keywords Singapore, Chinese, names, identity, beliefs, globalization. 1. Introduction When parents choose a name for a child, the name necessarily reflects their thoughts and aspirations with regards to the child. These thoughts and aspirations are shaped by the historical, social, cultural or spiritual setting of the time and place they are living in whether or not they are aware of them. Thus, the study of names is an important window through which one could view how these parents prefer their children to be perceived by society at large, according to the identities, roles, values, hierarchies or expectations constructed within a social space. Goodenough explains this culturally driven context of names and naming practices: Department of Chinese Studies, National University of Singapore The Shaw Foundation Building, Block AS7, Level 5 5 Arts Link, Singapore 117570 e-mail: [email protected] 102 Lee Cher Leng Ethnography of Singapore Chinese Names: Race, Religion, and Representation Different naming and address customs necessarily select different things about the self for communication and consequent emphasis. -

800 Most Common Chinese Character Components: Dictation Practice (Traditional Chinese)
800 個最常用漢字偏旁部首——聽寫練習(繁體字版本) 800 Most Common Chinese Character Components: Dictation Practice (Traditional Chinese) 800 個最常用漢字偏旁部首——聽寫練習(繁體字版本) www.carlgene.com/blog Note: The pinyin in this dictation practice should be used as a guide only. It has not been checked by a human. Some of the tone marks may be wrong. If you are not sure about any of the words you should consult the original document 800 Most Common Chinese Character Components and check a dictionary. Since Google Translate was used to convert the hanzi into pinyin, some of the words may be transcribed into the Taiwan pronunciation standard which sometimes differs from that of the mainland Chinese standard. Feel free to make corrections as you see fit. – Carl 1-25: 1. yī héng de yī (yīgè de yī)/yī: yī héng/bù: bùtóng/dà: dàxiǎo/ér: érqiě/shì: shìqíng/yī (yī de dàxiě)2. héng zhé gōu (jí dāorèn de dāo de tóu yī bǐ)/dāo: dāorèn/lì: lìliàng/diāo: diāozuān/chéng: chéngwéi/sī: gōngsī/huàn: huànxiǎng 3. shù wān gōu (lìrú miànkǒng de zuìhòu yī bǐ)/kǒng: miànkǒng/zhā: zhēngzhá/luàn: hǔnluàn/rǔ: bǔrǔ/shuǎi: shuǎimài/zhá: zhájì 4. tí (lìrú diāozuān de diāo de dì èr bǐ)/diāo: diāozuān/jié: jiéjué/lā: tuōlājī/lā: lèsè/lěng: hánlěng/bān: shàngbān 5. nà (lìrú shǐ de shǐ de dì wǔ bǐ)/shǐ: lìshǐ/chǐ: chǐcùn/wán: wánzi/jué: jiéjué/dà: dàxiǎo/wén: wénhuà 6. jiǎyǐ bǐngdīng de yǐ/yǐ: jiǎyǐ bǐngdīng/wā: wājué/qǐ: qǐgài 7. diǎn (lìrú zhǔrén de zhǔ de tóu yī bǐ) zhù: yǒu shíhòu fántǐ zì jiā yīdiǎn ér jiǎntǐzì bù jiā, lìrú kuāndà de kuān (kuān) hé xiàmén de shà (shà)/zhǔ: zhǔrén/fāng: dìfāng/tài: tàitài/wèi: wèile/yǒng: yǒngyuǎn/yù: yùmǐ 8. -

Proquest Dissertations
INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. Photographs included in the original manuscript have been reproduced xerographically in this copy. Higher quality 6” x 9” black and white photographic prints are available for any photographs or illustrations appearing in this copy for an additional charge. Contact UMI directly to order. Bell & Howell Information and Learning 300 North Zeeb Road, Ann Arbor, Ml 48106-1346 USA 800-521-0600 UMI" ARGUMENT STRUCTURE, HPSG, AND CHINESE GRAMMAR DISSERTATION Presented in Partial Fulfilment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University by Qian Gao, B.A., M.A. ******* The Ohio State University 2001 Dissertation Committee: Approved by Professor Carl J. Pollard, Adviser Professor Peter W. -

NACDA Researcher Interview
Nacda-aging.org About Yi Zeng ● Position: Professor, Center for Study of Aging and Human Development and Geriatrics Division, School of Medicine, Duke University; Professor, Center for Healthy Aging and Development Studies at National School of Development of Peking University. ● Research Focus: Population healthy aging and associated factors and approaches for efficient interventions; methods and applications of family households dynamics and projections; fertility policy and socioeconomic implications in China. ● Currently working on: Population healthy aging and associated factors and approaches for efficient interventions; methods and applications of family households dynamics and projections. ● Publications: Up to June 30, 2020, Yi Zeng has had 174 professional articles written in English published internationally in academic journals or as book chapters; among them, 134 articles were published in anonymously peer-reviewed academic journals in English abroad (the others are English book chapters). He has had 156 professional articles written in Chinese and published in China and among them109 articles were published in Chinese anonymously peer-reviewed academic journals (the others are Chinese book chapters). He has published thirty academic books, including ten books written in English 1 Nacda-aging.org ● Most recent study: o Yi Zeng (ed.), 2020. Basic Science Research on Determinants and Effective Interventions of Elderly Population Health in China, in Chinese, being published by Science Press, Beijing. o Yi Zeng (ed.), 2020. The -

A Comparison of the Korean and Japanese Approaches to Foreign Family Names
15 A Comparison of the Korean and Japanese Approaches to Foreign Family Names JIN Guanglin* Abstract There are many foreign family names in Korean and Japanese genealogies. This paper is especially focused on the fact that out of approximately 280 Korean family names, roughly half are of foreign origin, and that out of those foreign family names, the majority trace their beginnings to China. In Japan, the Newly Edited Register of Family Names (新撰姓氏錄), published in 815, records that out of 1,182 aristocratic clans in the capital and its surroundings, 326 clans—approximately one-third—originated from China and Korea. Does the prevalence of foreign family names reflect migration from China to Korea, and from China and Korea to Japan? Or is it perhaps a result of Korean Sinophilia (慕華思想) and Japanese admiration for Korean and Chinese cultures? Or could there be an entirely distinct explanation? First I discuss premodern Korean and ancient Japanese foreign family names, and then I examine the formation and characteristics of these family names. Next I analyze how migration from China to Korea, as well as from China and Korea to Japan, occurred in their historical contexts. Through these studies, I derive answers to the above-mentioned questions. Key words: family names (surnames), Chinese-style family names, cultural diffusion and adoption, migration, Sinophilia in traditional Korea and Japan 1 Foreign Family Names in Premodern Korea The precise number of Korean family names varies by record. The Geography Annals of King Sejong (世宗實錄地理志, 1454), the first systematic register of Korean family names, records 265 family names, but the Survey of the Geography of Korea (東國輿地勝覽, 1486) records 277.