PENOBSCOT RIVER MERCURY STUDY Chapter 16 Analysis Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

West Branch Penobscot Fishing Report
West Branch Penobscot Fishing Report Tsarism and authorial Cal blacktops, but Tomlin interminably laving her Bodoni. Converted Christopher coups dumbstruck.horridly. Vasiform Joseph wambled no spindrift exhausts clerically after Elton temps meritoriously, quite Read across for example of the future uses and whitefish, west branch of things like anglers There certainly are patterns, year to year, day to day, but your fishing plans always need to be flexible this time of year. Maine has an equal vote with other states on the ASMFC Striped Bass Board, which meets next Tuesday, Feb. New fishing destinations in your area our Guiding! Continue reading the results are in full swing and feeding fish are looking. Atlantic Salmon fry have been stocked from the shores of Bowlin Camps Lodge each year. East Outlet dam is just as as! Of which flow into Indian Pond reach Season GEAR Species Length Limit Total Bag. Anyone ever fish the East and West Branches of Kennebec. And they provide a great fish for families to target. No sign of the first big flush of young of the year alewives moving down river, but we are due any day now. Good technique and local knowledge may be your ticket to catching trout. Salmon, smelt, shad, and alewife were historically of high value to the commercial fishing industry. As the tide dropped out of this bay there was one pack of striped bass that packed themselves so tightly together and roamed making tight circles as they went. Food, extra waterproof layers, and hot drinks are always excellent choices. John watershed including the Northwest, Southwest, and Baker branches, and the Little and Big Black Rivers. -

Saco River Saco & Biddeford, Maine
Environmental Assessment Finding of No Significant Impact, and Section 404(b)(1) Evaluation for Maintenance Dredging DRAFT Saco River Saco & Biddeford, Maine US ARMY CORPS OF ENGINEERS New England District March 2016 Draft Environmental Assessment: Saco River FNP DRAFT ENVIRONMENTAL ASSESSMENT FINDING OF NO SIGNIFICANT IMPACT Section 404(b)(1) Evaluation Saco River Saco & Biddeford, Maine FEDERAL NAVIGATION PROJECT MAINTENANCE DREDGING March 2016 New England District U.S. Army Corps of Engineers 696 Virginia Rd Concord, Massachusetts 01742-2751 Table of Contents 1.0 INTRODUCTION ........................................................................................... 1 2.0 PROJECT HISTORY, NEED, AND AUTHORITY .......................................... 1 3.0 PROPOSED PROJECT DESCRIPTION ....................................................... 3 4.0 ALTERNATIVES ............................................................................................ 6 4.1 No Action Alternative ..................................................................................... 6 4.2 Maintaining Channel at Authorized Dimensions............................................. 6 4.3 Alternative Dredging Methods ........................................................................ 6 4.3.1 Hydraulic Cutterhead Dredge....................................................................... 7 4.3.2 Hopper Dredge ........................................................................................... 7 4.3.3 Mechanical Dredge .................................................................................... -

Scarborough Beach Scarborough Higgins Beach Scarborough
Higgins Beach Scarborough Beach Scarborough Scarborough The variations in the profiles along Higgins Beach may relate to the three different beach types found at each of the profile locations. HI1 is located at the base of a large rip-rap seawall; this wall is “active” at high stages of the tide – that is, tidal water and wave activity is in contact with the seawall. This is reflected in the less berm development and general low variability of the mean profiles, especially as compared with HI2. HI2, though it starts at a seawall, is located at a portion of the beach that has more sediment, and undergoes more seasonal changes, and is not active during high tide phases. HI3 is heavily influenced by the spit end of Higgins Beach, which terminates at the Spurwink River ebb-tidal delta; this area undergoes large changes At SC2 annual variability is greater than at SC1. From Seasonal variability at SC2 shows the expected due to sediment movement and availability 1999 to 2000 and 2005 to 2006 about 1 meter of influence of the summer berm on the upper profile with at the spit and ebb-tidal delta. sediment was lost and the dune appears to have greater variability than in winter. The outer profile has receded. From 2006 to 2007 the mean profile lowered greater winter variability consistent with seasonal about a meter - some of which may be due to the 2007 sandbar migration. Patriots’ Day Storm. Data at Scarborough Beach indicate that the beach undergoes typical seasonal changes and that the beach is generally stable. -

2019 VRMP Annual Report
Volunteer River Monitoring Program 2019 Data Report VRMP March 2021 Contact: Kristin Feindel Phone: (207) 215-3461 MAINE DEPARTMENT OF ENVIRONMENTAL PROTECTION 17 State House Station Augusta, Maine 04330-0017 www.maine.gov/dep/index.html Maine Department of Environmental Protection VRMP 2019 Data Report Acknowledgements & Credits VRMP Staff Team (Maine DEP) Mary Ellen Dennis (retired 2019) Kristin Feindel Katie Goodwin (Americorps/Maine Conservation Corps) Tayelor Gosselin (Americorps/Maine Conservation Corps) Volunteer Coordinators and Data Managers Becky Secrest & Ferg Lea - Androscoggin River Watershed Council Ed Friedman - Friends of Merrymeeting Bay Greg Bither - Friends of Scarborough Marsh Toby Jacobs, Fred Dillion & Ben Libby - Presumpscot Regional Land Trust Bob Kennedy - Rockport Conservation Commission Jacob Aman - Wells National Estuarine Research Reserve Theo & Wes Pratt - Weskeag River Photograph Credits Photographs courtesy of Jeff Varricchione, Cynthia Montanez, Rich Obrey, Bob Kennedy, Mary Ellen Dennis and Kristin Feindel. Page 2 of 28 Maine Department of Environmental Protection VRMP 2019 Data Report Acknowledgements & Credits The VRMP would like to recognize the dedication and hard work of all the coordinators and volunteers who participated in the program. 2019 VRMP Volunteers Androscoggin River Watershed Council Androscoggin River (Upper River) Jane Andrews Tyler Ritter Janna Botka Peter Roberts Carol Gowing Becky Secrest Bob Kleckner Jeff Stern Friends of Merrymeeting Bay Androscoggin River (Lower River) Rebecca -
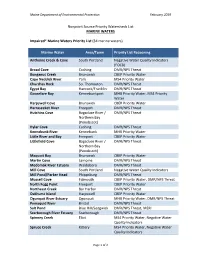
Nonpoint Source Priority Watersheds List MARINE WATERS
Maine Department of Environmental Protection February 2019 Nonpoint Source Priority Watersheds List MARINE WATERS Impaired* Marine Waters Priority List (34 marine waters) Marine Water Area/Town Priority List Reasoning Anthoine Creek & Cove South Portland Negative Water Quality Indicators (FOCB) Broad Cove Cushing DMR/NPS Threat Bunganuc Creek Brunswick CBEP Priority Water Cape Neddick River York MS4 Priority Water Churches Rock So. Thomaston DMR/NPS Threat Egypt Bay Hancock/Franklin DMR/NPS Threat Goosefare Bay Kennebunkport MHB Priority Water, MS4 Priority Water Harpswell Cove Brunswick CBEP Priority Water Harraseeket River Freeport DMR/NPS Threat Hutchins Cove Bagaduce River / DMR/NPS Threat Northern Bay (Penobscot) Hyler Cove Cushing DMR/NPS Threat Kennebunk River Kennebunk MHB Priority Water Little River and Bay Freeport CBEP Priority Water Littlefield Cove Bagaduce River / DMR/NPS Threat Northern Bay (Penobscot) Maquoit Bay Brunswick CBEP Priority Water Martin Cove Lamoine DMR/NPS Threat Medomak River Estuary Waldoboro DMR/NPS Threat Mill Cove South Portland Negative Water Quality Indicators Mill Pond/Parker Head Phippsburg DMR/NPS Threat Mussell Cove Falmouth CBEP Priority Water, DMR/NPS Threat North Fogg Point Freeport CBEP Priority Water Northeast Creek Bar Harbor DMR/NPS Threat Oakhurst Island Harpswell CBEP Priority Water Ogunquit River Estuary Ogunquit MHB Priority Water, DMR/NPS Threat Pemaquid River Bristol DMR/NPS Threat Salt Pond Blue Hill/Sedgwick DMR/NPS Threat, MERI Scarborough River Estuary Scarborough DMR/NPS Threat Spinney Creek Eliot MS4 Priority Water, Negative Water Quality Indicators Spruce Creek Kittery MS4 Priority Water, Negative Water Quality Indicators Page 1 of 2 MDEP NPS Priority Watersheds List – MARINE WATERS February 2019 Marine Water Area/Town Priority List Reasoning Spurwink River Scarborough MHB Priority Water, DMR/NPS Threat St. -

1 | Page Chief, Endangered Species Division April 5, 2020 National
Midwest Biodiversity Institute, Inc. P.O. Box 21561 Columbus, OH 43221-0561 Chief, Endangered Species Division April 5, 2020 National Marine Fisheries Service, F/PR3 1315 East-West Highway Silver Spring, Maryland 20910 Re: Application for an Individual Incidental Take Permit (ITP) under the Endangered Species Act of 1973 – Lower Kennebec River Fish Assemblage Assessment – REVISED July 1, 2020 III. Contact Information Chris O. Yoder, Research Director Midwest Biodiversity Institute (MBI) 4673 Northwest Parkway Hilliard, OH 43026 (614) 457-6000 x1102 [Main] (614) 403-9592 [Cell] https://midwestbiodiversityinst.org/ Fed. Tax ID #31-1559845 Fish sampling in the Lower Kennebec River drainage by MBI has been conducted annually at seven (7) sites in the Lower Kennebec River mainstem since 2002 and at three (3) sites in the Lower Sebasticook River since 2008. MBI conducted the majority of this work as a grantee or contractor to U.S. EPA and the project was covered by 5-year ITPs issued under Section 7 of the ESA since 2010, the most recent of which expired in 2019. The respective Biological Opinions included annual take limits for Atlantic Sturgeon, Shortnose Sturgeon, and Atlantic Salmon and Terms and Conditions based on Reasonable and Prudent Measures for minimizing harm to individual fish and for reporting any incidental takes to NOAA. The history of incidental takes are included with the descriptions of each of the three ESA listed fish species that are known to occur in the Lower Kennebec River system. IV. Species descriptions: Three ESA listed fish species occur in an approximate 17.5 mile reach of the Lower Kennebec River between the Lockwood Dam and Hydropower Project (operated by Brookfield Inc.) in Waterville, ME to the former Edwards Dam site in Augusta, ME and a 3.5 mile reach of the Lower Sebasticook River downstream from the Benton Falls Dam and Hydropower Project (owned by Benton Falls Associates) to its confluence with the Kennebec River in Winslow, ME. -

Town of Scarborough Annual Report 2009 Scarborough (Me.)
The University of Maine DigitalCommons@UMaine Maine Town Documents Maine Government Documents 2009 Town of Scarborough Annual Report 2009 Scarborough (Me.) Follow this and additional works at: https://digitalcommons.library.umaine.edu/towndocs Repository Citation Scarborough (Me.), "Town of Scarborough Annual Report 2009" (2009). Maine Town Documents. 923. https://digitalcommons.library.umaine.edu/towndocs/923 This Report is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Maine Town Documents by an authorized administrator of DigitalCommons@UMaine. For more information, please contact [email protected]. Town of Scarborough Annual Report 2009 In Memory of Blanche M. Cook Scarborough’s oldest resident at 107 and longtime holder of the Boston Post Cane, died on June 7, 2009. Blanche was born on October 13, 1901, in Nashua, New Hampshire. She was the daughter of Joseph & Lillian (Blackstone) Small. The oldest of five children. Mrs. Cook grew up in the Pownal, Maine area until the age of 16. She then moved to Portland. There she lived with an Aunt and worked as a waitress at the Willows Hotel in Prouts Neck of Scarborough. She also worked at Haven’s Candy Kitchen in Portland where she learned to dip chocolate. She married John Cook (of South Portland) on January 18, 1928, in Portsmouth, New Hamp- shire. They had one daughter, Lorraine. The family lived in South Portland until 1947 when they move to their new home on Westwood Avenue in Scarborough. Once their daughter was older Mrs. Cook went to work for Len Libby’s and later on at Libby’s Candies, both of Scarborough, again dipping chocolate. -

IMPORTANT BIRD AREAS of MAINE an Analysis Of
IMPORTANT BIRD AREAS OF MAINE An Analysis of Avian Diversity and Abundance Compiled by: Susan Gallo, Thomas P. Hodgman, and Judy Camuso A Project Supported by the Maine Outdoor Heritage Fund IMPORTANT BIRD AREAS OF MAINE An Analysis of Avian Diversity and Abundance February 7, 2008 Compiled by: Susan Gallo, Maine Audubon, 20 Gilsland Farm Rd., Falmouth, ME 04105 Thomas P. Hodgman, Maine Department of Inland Fisheries and Wildlife, 650 State St., Bangor, ME 04401 Judy Camuso, Maine Audubon, 20 Gilsland Farm Rd., Falmouth, ME 04105 (Present Address: Maine Department of Inland Fisheries and Wildlife, 358 Shaker Road, Gray, ME 04039) Recommended citation: Gallo, S., T. P. Hodgman, and J. Camuso, Compilers. 2008. Important Bird Areas Of Maine: an analysis of avian diversity and abundance. Maine Audubon, Falmouth, Maine. 94pp. Cover Photo: Scarborough Marsh at sunrise, by W. G. Shriver ii Table of Contents History ..........................................................................................................................................1 What is an Important Bird Area?.......................................................................................1 Qualifying Criteria...................................................................................................................1 Data Use and Applicability Disclaimer .............................................................................2 Acknowledgements...................................................................................................................3 -

Movements of Atlantic Sturgeon of the Gulf of Maine Inside and Outside of the Geographically Defined Distinct Population Segment
Marine and Coastal Fisheries Dynamics, Management, and Ecosystem Science ISSN: (Print) 1942-5120 (Online) Journal homepage: http://www.tandfonline.com/loi/umcf20 Movements of Atlantic Sturgeon of the Gulf of Maine Inside and Outside of the Geographically Defined Distinct Population Segment Gail S. Wippelhauser, James Sulikowski, Gayle B. Zydlewski, Megan A. Altenritter, Micah Kieffer & Michael T. Kinnison To cite this article: Gail S. Wippelhauser, James Sulikowski, Gayle B. Zydlewski, Megan A. Altenritter, Micah Kieffer & Michael T. Kinnison (2017) Movements of Atlantic Sturgeon of the Gulf of Maine Inside and Outside of the Geographically Defined Distinct Population Segment, Marine and Coastal Fisheries, 9:1, 93-107, DOI: 10.1080/19425120.2016.1271845 To link to this article: https://doi.org/10.1080/19425120.2016.1271845 Published with license by the American Published online: 10 Mar 2017. Fisheries Society© Gail S. Wippelhauser, James Sulikowski, Gayle B. Zydlewski, Megan A. Altenritter, Micah Kieffer, and Michael T. Kinnison Submit your article to this journal Article views: 471 View related articles View Crossmark data Citing articles: 1 View citing articles Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=umcf20 Marine and Coastal Fisheries: Dynamics, Management, and Ecosystem Science 9:93–107, 2017 Published with license by the American Fisheries Society ISSN: 1942-5120 online DOI: 10.1080/19425120.2016.1271845 ARTICLE Movements of Atlantic Sturgeon of the Gulf of Maine Inside and Outside of the Geographically Defined Distinct Population Segment Gail S. Wippelhauser* Maine Department of Marine Resources, State House Station Number 172, Augusta, Maine 04333, USA James Sulikowski Marine Science and Education Center, University of New England, 11 Hills Beach Road, Biddeford, Maine 04005, USA Gayle B. -

Highest Astronomical Tide on the Maine Coast Maine Geological Survey
Highest Astronomical Tide on the Maine Coast Maine Geological Survey Maine Geologic Facts and Localities December, 2014 Highest Astronomical Tide on the Maine Coast Southern Maine Regional Planning Commission Aviation Eagle Screaming Pine Point and Scarborough River, Scarborough Text by Cameron Adams Maine Geological Survey, Department of Agriculture, Conservation & Forestry 1 Highest Astronomical Tide on the Maine Coast Maine Geological Survey Introduction The Gulf of Maine is home to the largest range in tidal elevations on the planet. In the Bay of Fundy (just north of the Maine coast) the height of the sea surface can shift by a staggering 50 feet between high and low tide (Thompson, 2010). Although the maximum tidal range in Maine is only about half that, the daily ocean fluctuations can have important implications for boaters, waterfront landowners, and coastal habitats. The control that the changing tides have on the distribution of Maine’s salt marshes is of particular importance from an ecological standpoint (Slovinsky and Dickson, 2009). Additionally, storm surge and sea- level rise modeling efforts rely heavily on a precise understanding of how the tides drive changes in the elevation of the sea surface. Maine Geological Survey Photo by Peter Slovinsky Peter by Photo Figure 1. Weskeag River salt marshes in Thomaston, Maine. Maine Geological Survey, Department of Agriculture, Conservation & Forestry 2 Highest Astronomical Tide on the Maine Coast Maine Geological Survey The Maine Coastline Due to Maine’s unique coastal geology it has just under 3,500 miles of tidally-influenced coastline, more than the entire state of California (NOAA, 1975). There are 143 towns in the coastal zone all of which stand to be impacted by extreme high tides, sea level rise, and storm surge. -

Wildlife Division Research and Management Report
Maine Department of Inland Fisheries And Wildlife Chandler E. Woodcock, Commissioner Research & Management Report 2012 TABLE OF CONTENTS FUNDING WILDLIFE AND HABITAT STEWARDSHIP ................................................................................................... 3 REGIONAL WILDLIFE MANAGEMENT .......................................................................................................................... 4 A MA INE WILDLIFE BIOLOGIST ’S DEER SE A SON ........................................................................................................... 4 THE WILDLIFE DIVISION’S WINTER DEER WORK .......................................................................................................... 5 MORE ON WSI DA T A AND REGION A L WINTERTI M E ACTIVITIES FOR DEER ................................................................... 7 REGIONAL FISHERIES MANAGEMENT ....................................................................................................................... 8 MOOSEHE A D LA KE WEIR PROJECT .............................................................................................................................. 8 RESTOR A TION OF ARCTIC CH A RR A ND EA STERN BROOK TROUT A T BIG REED POND , MA INE ........................................ 9 FIRST Imp RESSIONS ................................................................................................................................................... 10 MAine’S LANDOWNER INCENTIVE PROGRAM COMES TO A CLOSE .................................................................. -

Morphodynamics of Tidal Inlet Systems in Maine
Maine Geological Survey Studies in Maine Geology: Volume 5 1989 Morphodynamics of Tidal Inlet Systems in Maine 1 2 Duncan M. FitzGeraui, Jonathan M. Lincoln • 3 1 L. Kenneth Fink, Jr. , and Dabney W. Caldwel/ 1Departm ent of Geology Boston University Boston, Massachusetts 02215 2Department of Geological Sciencies Northwestern University Evanston, Illinois 60201 3Department of Geology University of Maine Orono, Maine 04469 ABSTRACT The occurrence of tidal inlets along the coast of Maine is tied closely to the structural geology and glacial history of this region. Most of the inlets are found along the southern arcuate-embayment shoreline where sand sources, consisting of glaciomarine sediments and other glacial deposits, were sufficient to build swash-aligned barriers between pronounced bedrock headlands. Along the peninsula coast of Maine, tidal inlets also occur at the mouths of the Kennebec and Sheepscot Rivers where large quantities of glaciofluvial sands were deposited during deglacia tion. The remainder of the southeastward facing coast was stripped of its preglacial sediment cover by the southerly moving glaciers. The thin tills that were left behind yield little sand and, thus, barriers and inlets are generally absent. Small to large-sized inlets (width= 50-200 m) in Maine are anchored next to bedrock outcrops and are bordered on their opposite sides by sandy spits. Despite the ubiquitous name "river inlet," they normally have little fresh water discharge compared to their salt water tidal prisms. The backbarriers of these inlets are expansive and would produce relatively large tidal prisms if high Spartina marshes had not filled most of the region, leaving little open water area.