Enhanced Antibacterial Activity of Ent-Labdane Derivatives Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Relative Rates of Thiol–Thioester Exchange and Hydrolysis for Alkyl and Aryl Thioalkanoates in Water
Orig Life Evol Biosph (2011) 41:399–412 DOI 10.1007/s11084-011-9243-4 PREBIOTIC CHEMISTRY The Relative Rates of Thiol–Thioester Exchange and Hydrolysis for Alkyl and Aryl Thioalkanoates in Water Paul J. Bracher & Phillip W. Snyder & Brooks R. Bohall & George M. Whitesides Received: 14 April 2011 /Accepted: 16 June 2011 / Published online: 5 July 2011 # Springer Science+Business Media B.V. 2011 Abstract This article reports rate constants for thiol–thioester exchange (kex), and for acid- mediated (ka), base-mediated (kb), and pH-independent (kw) hydrolysis of S-methyl thioacetate and S-phenyl 5-dimethylamino-5-oxo-thiopentanoate—model alkyl and aryl thioalkanoates, respectively—in water. Reactions such as thiol–thioester exchange or aminolysis could have generated molecular complexity on early Earth, but for thioesters to have played important roles in the origin of life, constructive reactions would have needed to compete effectively with hydrolysis under prebiotic conditions. Knowledge of the kinetics of competition between exchange and hydrolysis is also useful in the optimization of systems where exchange is used in applications such as self-assembly or reversible binding. For the alkyl thioester S-methyl thioacetate, which has been synthesized in −5 −1 −1 −1 −1 −1 simulated prebiotic hydrothermal vents, ka = 1.5×10 M s , kb = 1.6×10 M s , and −8 −1 kw = 3.6×10 s . At pH 7 and 23°C, the half-life for hydrolysis is 155 days. The second- order rate constant for thiol–thioester exchange between S-methyl thioacetate and 2- −1 −1 sulfonatoethanethiolate is kex = 1.7 M s . -

Chapter 7, Haloalkanes, Properties and Substitution Reactions of Haloalkanes Table of Contents 1
Chapter 7, Haloalkanes, Properties and Substitution Reactions of Haloalkanes Table of Contents 1. Alkyl Halides (Haloalkane) 2. Nucleophilic Substitution Reactions (SNX, X=1 or 2) 3. Nucleophiles (Acid-Base Chemistry, pka) 4. Leaving Groups (Acid-Base Chemistry, pka) 5. Kinetics of a Nucleophilic Substitution Reaction: An SN2 Reaction 6. A Mechanism for the SN2 Reaction 7. The Stereochemistry of SN2 Reactions 8. A Mechanism for the SN1 Reaction 9. Carbocations, SN1, E1 10. Stereochemistry of SN1 Reactions 11. Factors Affecting the Rates of SN1 and SN2 Reactions 12. --Eliminations, E1 and E2 13. E2 and E1 mechanisms and product predictions In this chapter we will consider: What groups can be replaced (i.e., substituted) or eliminated The various mechanisms by which such processes occur The conditions that can promote such reactions Alkyl Halides (Haloalkane) An alkyl halide has a halogen atom bonded to an sp3-hybridized (tetrahedral) carbon atom The carbon–chlorine and carbon– bromine bonds are polarized because the halogen is more electronegative than carbon The carbon-iodine bond do not have a per- manent dipole, the bond is easily polarizable Iodine is a good leaving group due to its polarizability, i.e. its ability to stabilize a charge due to its large atomic size Generally, a carbon-halogen bond is polar with a partial positive () charge on the carbon and partial negative () charge on the halogen C X X = Cl, Br, I Different Types of Organic Halides Alkyl halides (haloalkanes) sp3-hybridized Attached to Attached to Attached -

Nomenclature of Carboxylic Acids • Select the Longest Carbon Chain Containing the Carboxyl Group
Chapter 5 Carboxylic Acids and Esters Carboxylic Acids • Carboxylic acids are weak organic acids which Chapter 5 contain the carboxyl group (RCO2H): Carboxylic Acids and Esters O C O H O RCOOH RCO2H Chapter Objectives: O condensed ways of • Learn to recognize the carboxylic acid, ester, and related functional groups. RCOH writing the carboxyl • Learn the IUPAC system for naming carboxylic acids and esters. group a carboxylic acid C H • Learn the important physical properties of the carboxylic acids and esters. • Learn the major chemical reaction of carboxylic acids and esters, and learn how to O predict the products of ester synthesis and hydrolysis reactions. the carboxyl group • Learn some of the important properties of condensation polymers, especially the polyesters. Mr. Kevin A. Boudreaux • The tart flavor of sour-tasting foods is often caused Angelo State University CHEM 2353 Fundamentals of Organic Chemistry by the presence of carboxylic acids. Organic and Biochemistry for Today (Seager & Slabaugh) www.angelo.edu/faculty/kboudrea 2 Nomenclature of Carboxylic Acids • Select the longest carbon chain containing the carboxyl group. The -e ending of the parent alkane name is replaced by the suffix -oic acid. • The carboxyl carbon is always numbered “1” but the number is not included in the name. • Name the substituents attached to the chain in the Nomenclature of usual way. • Aromatic carboxylic acids (i.e., with a CO2H Carboxylic Acids directly connected to a benzene ring) are named after the parent compound, benzoic acid. O C OH 3 -

Introduction to Alkenes and Alkynes in an Alkane, All Covalent Bonds
Introduction to Alkenes and Alkynes In an alkane, all covalent bonds between carbon were σ (σ bonds are defined as bonds where the electron density is symmetric about the internuclear axis) In an alkene, however, only three σ bonds are formed from the alkene carbon -the carbon thus adopts an sp2 hybridization Ethene (common name ethylene) has a molecular formula of CH2CH2 Each carbon is sp2 hybridized with a σ bond to two hydrogens and the other carbon Hybridized orbital allows stronger bonds due to more overlap H H C C H H Structure of Ethylene In addition to the σ framework of ethylene, each carbon has an atomic p orbital not used in hybridization The two p orbitals (each with one electron) overlap to form a π bond (p bonds are not symmetric about the internuclear axis) π bonds are not as strong as σ bonds (in ethylene, the σ bond is ~90 Kcal/mol and the π bond is ~66 Kcal/mol) Thus while σ bonds are stable and very few reactions occur with C-C bonds, π bonds are much more reactive and many reactions occur with C=C π bonds Nomenclature of Alkenes August Wilhelm Hofmann’s attempt for systematic hydrocarbon nomenclature (1866) Attempted to use a systematic name by naming all possible structures with 4 carbons Quartane a alkane C4H10 Quartyl C4H9 Quartene e alkene C4H8 Quartenyl C4H7 Quartine i alkine → alkyne C4H6 Quartinyl C4H5 Quartone o C4H4 Quartonyl C4H3 Quartune u C4H2 Quartunyl C4H1 Wanted to use Quart from the Latin for 4 – this method was not embraced and BUT has remained Used English order of vowels, however, to name the groups -

Linear Alkylbenzene Sulphonate (CAS No
Environmental Risk Assessment LAS Linear Alkylbenzene Sulphonate (CAS No. 68411-30-3) Revised ENVIRONMENTAL Aspect of the HERA Report = February 2013 = 1 1. Contents 2. Executive summary 3. Substance characterisation 3.1 CAS No. and grouping information 3.2 Chemical structure and composition 3.3 Manufacturing route and production/volume statistics 3.4 Consumption scenario in Europe 3.5 Use application summary 4. Environmental safety assessment 4.1 Environmental exposure assessment 4.1.1 Biotic and abiotic degradability 4.1.2 Removal 4.1.3 Monitoring studies 4.1.4 Exposure assessment: scenario description 4.1.5 Substance data used for the exposure calculation 4.1.6 PEC calculations 4.1.7 Bioconcentration 4.2 Environmental effects assessment 4.2.1 Ecotoxicity 4.2.1.1 Aquatic ecotoxicity 4.2.1.2 Terrestrial ecotoxicity 4.2.1.3 Sediment ecotoxicity 4.2.1.4 Ecotoxicity to sewage microorganisms 4.2.1.5 Reassurance on absence of estrogenic effects 4.2.2 PNEC calculations 4.2.2.1 Aquatic PNEC 4.2.2.2 Terrestrial PNEC 4.2.2.3 Sludge PNEC 4.2.2.4 Sediment PNEC 4.2.2.5 STP PNEC 4.3 Environment risk assessment 5. 5. References 6. Contributors to the report 6.1 Substance team 6.2 HERA environmental task force 6.3 HERA human health task force 6.4 Industry coalition for the OECD/ICCA SIDS assessment of LAS 2 2. Executive Summary Linear alkylbenzene sulphonate (LAS) is an anionic surfactant. It was introduced in 1964 as the readily biodegradable replacement for highly branched alkylbenzene sulphonates (ABS). -

Chapter 7 - Alkenes and Alkynes I
Andrew Rosen Chapter 7 - Alkenes and Alkynes I 7.1 - Introduction - The simplest member of the alkenes has the common name of ethylene while the simplest member of the alkyne family has the common name of acetylene 7.2 - The (E)-(Z) System for Designating Alkene Diastereomers - To determine E or Z, look at the two groups attached to one carbon atom of the double bond and decide which has higher priority. Then, repeat this at the other carbon atom. This system is not used for cycloalkenes - If the two groups of higher priority are on the same side of the double bond, the alkene is designated Z. If the two groups of higher priority are on opposite sides of the double bond, the alkene is designated E - Hydrogenation is a syn/cis addition 7.3 - Relative Stabilities of Alkenes - The trans isomer is generally more stable than the cis isomer due to electron repulsions - The addition of hydrogen to an alkene, hydrogenation, is exothermic (heat of hydrogenation) - The greater number of attached alkyl groups, the greater the stability of an alkene 7.5 - Synthesis of Alkenes via Elimination Reactions, 7.6 - Dehydrohalogenation of Alkyl Halides How to Favor E2: - Reaction conditions that favor elimination by E1 should be avoided due to the highly competitive SN 1 mechanism - To favor E2, a secondary or tertiary alkyl halide should be used - If there is only a possibility for a primary alkyl halide, use a bulky base - Use a higher concentration of a strong and nonpolarizable base, like an alkoxide - EtONa=EtOH favors the more substituted double bond -
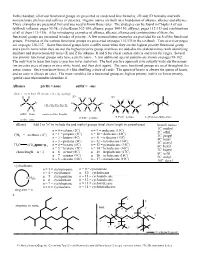
In This Handout, All of Our Functional Groups Are Presented As Condensed Line Formulas, 2D and 3D Formulas and with Nomenclature Prefixes and Suffixes (If Present)
In this handout, all of our functional groups are presented as condensed line formulas, 2D and 3D formulas and with nomenclature prefixes and suffixes (if present). Organic names are built on a foundation of alkanes, alkenes and alkynes. Those examples are presented first and you need to know those rules. The strategies can be found in Chapter 4 of our textbook (alkanes: pages 93-98, cycloalkanes 102-104, alkenes: pages 104-110, alkynes: pages 112-113 and combinations of all of them 113-115). After introducing examples of alkanes, alkenes, alkynes and combinations of them, the functional groups are presented in order of priority. A few nomenclature examples are provided for each of the functional groups. Examples of the various functional groups are presented on pages 115-135 in the textbook. Two overview pages are on pages 136-137. Some functional groups have a suffix name when they are the highest priority functional group and a prefix name when they are not the highest priority group, and these are added to the skeletal names with identifying numbers and stereochemistry terms (E and Z for alkenes, R and S for chiral centers and cis and trans for rings). Several low priority functional groups only have a prefix name. A few additional special patterns are shown on pages 98-102. The only way to learn this topic is practice (over and over). The best practice approach is to actually write out the names (on an extra piece of paper or on a white board, and then do it again). The same functional groups are used throughout the entire course. -

Nomenclature and Conformations of Alkanes and Cycloalkanes1 on The
Andrew Rosen Chapter 4 - Nomenclature and Conformations of Alkanes and Cycloalkanes1 4.1 - Introduction to Alkanes and Cycloalkanes - Alkanes are hydrocarbons with all carbon-carbon single bonds - Alkenes are hydrocarbons with a carbon-carbon double bond - Alkynes are hydrocarbons with a carbon-carbon triple bond - Cycloalkanes are alkanes in which all or some of the carbon atoms are arranged in a ring - Alkanes have the general formula of CnH2n+2 - Cycloalkanes with a single ring have the formula CnH2n 4.2 - Shapes of Alkanes - An unbranched chain means that each carbon atom is bonded to no more than two other carbon atoms and that there are only primary and secondary carbon atoms 4.3 - IUPAC Nomenclature of Alkanes, Alkyl Halides, and Alcohols - Endings of alkane names end in −ane, and the standard prexes apply to how many carbon atoms there are (meth-, eth-, prop-, but-, pent-, etc.) - An alkyl group has one hydrogen removed from an alkane, and the names have a sux of −yl Rules for naming branched-chain alkanes: 1) Locate the longest continuous chain of carbon atoms. This chain determines the parent (prex) name for the alkane. Always start numbering from the end of a chain 2) Number the longest chain begining with the end of the chain nearer the substitutent 3) Use the numbers obtained by application of Rule 2 to designate the location of the substitutent group 4) When two or more substitutents are present, give each substituent a number corresponding to its location on the longest chain. Name them in alphabetical order 5) When two -

II. Nomenclature Rules for Alkenes 1. the Parent Name Will Be the Longest
1 Lecture 9 II. Nomenclature Rules For Alkenes 1. The parent name will be the longest carbon chain that contains both carbons of the double bond. Drop the -ane suffix of the alkane name and add the –ene suffix. Never name the double bond as a prefix. If a double bond is present, you have an alkene, not an alkane. alkane + -ene = alkene 2. Begin numbering the chain at the end nearest the double bond. Always number through the double bond and identify its position in the longest chain with the lower number. In the older IUPAC rules the number for the double bond was placed in front of the stem name with a hyphen. Under the newer rules, the number for the double bond is placed right in front of “ene”, with hyphens. We will use the newer rules for specifying the location of pi bonds. 1 2 3456 H3CCHCH CH 2 CH2 CH3 hex-2-ene (newer rules) 2-hexene (older rules) 3. Indicate the position of any substituent group by the number of the carbon atom in the parent (longest) chain to which it is attached. CH 1 2 345 3 H3CCHCHCHCH2 CH CH3 6 7 CH3 Numbering is determined by the double bond, not the branches, because the double bond has 5,6-dimethylhept-3-ene (newer rules) higher priority than any alkyl branch. 5,6-dimethyl-3-heptene (older rules) 4. Number cycloalkenes so that the double bond is 1,2 (number through the double bond). Number in the direction about the ring so that the lowest number is used at the first point of difference. -

Carboxylic Acids
CARBOXYLIC ACIDS 1 Carboxylic Acids Introduction Carboxylic acids are organic compounds containing the carboxyl group (-COOH), wherein the hydroxyl group (-OH) is directly attached to the carbonyl (C=O) group. Carboxylic acids constitute one of the most frequently encountered classes of organic compounds in nature. 2 Natural Carboxylic Acids A great many carboxylic acids are encountered in nature, mostly, in fruits. Indeed carboxylic acids were among the first class of organic compounds to ever be isolated from nature. Edible carboxylic acids found in citrous fruits and fermented milk generally have sharp flavours. 3 Nomenclature of Carboxylic Acids The common names of some basic carboxylic acids are derived from Latin names that indicate the first original natural source of the carboxylic acid. Structure of Acid Natural Source Common Name O H C OH Ants (Formica) Formic acid O CH3 C OH Vinegar (Acetum) Acetic acid O CH3CH2 C OH Basic Fat (Propio) Propionic acid O CH3CH2CH2 C OH Rancid butter (Butyrum) Butyric acid Present in aValerian herb Valeric acid O 4 CH3CH2CH2CH2CH2 C OH Goat (Caper) Caproic acid Common Names of Carboxylic Acids The common name of a carboxylic acid (R-COOH) is derived by adding the suffix –ic acid to a prefix representing the chain length of the carboxylic acid. # of Carbons Prefix Common Name of Acid 1 Form- Formic acid 2 Acet- Acetic acid 3 Propion- Propionic acid 4 Butyr- Butyric acid 5 Valer- Valeric acid 6 Capro- Caproic acid Aromatic acid Benzo- Benzoic acid 5 IUPAC Nomenclature of Aliphatic Carboxylic Acids IUPAC names of straight chain aliphatic carboxylic acids are derived by adding the suffix –oic acid to the systematic name of the parent hydrocarbon. -

Reactions of Alkenes Since Bonds Are Stronger Than Bonds, Double Bonds Tend to React to Convert the Double Bond Into Bonds
Reactions of Alkenes Since bonds are stronger than bonds, double bonds tend to react to convert the double bond into bonds This is an addition reaction. (Other types of reaction have been substitution and elimination). Addition reactions are typically exothermic. Electrophilic Addition The bond is localized above and below the C-C bond. The electrons are relatively far away from the nuclei and are therefore loosely bound. An electrophile will attract those electrons, and can pull them away to form a new bond. This leaves one carbon with only 3 bonds and a +ve charge (carbocation). The double bond acts as a nucleophile (attacks the electrophile). Ch08 Reacns of Alkenes (landscape) Page 1 In most cases, the cation produced will react with another nucleophile to produce the final overall electrophilic addition product. Electrophilic addition is probably the most common reaction of alkenes. Consider the electrophilic addition of H-Br to but-2-ene: The alkene abstracts a proton from the HBr, and a carbocation and bromide ion are generated. The bromide ion quickly attacks the cationic center and yields the final product. In the final product, H-Br has been added across the double bond. Ch08 Reacns of Alkenes (landscape) Page 2 Orientation of Addition Consider the addition of H-Br to 2-methylbut-2-ene: There are two possible products arising from the two different ways of adding H-Br across the double bond. But only one is observed. The observed product is the one resulting from the more stable carbocation intermediate. Tertiary carbocations are more stable than secondary. -

1 Chapter 3: Organic Compounds: Alkanes and Cycloalkanes
Chapter 3: Organic Compounds: Alkanes and Cycloalkanes >11 million organic compounds which are classified into families according to structure and reactivity Functional Group (FG): group of atoms which are part of a large molecule that have characteristic chemical behavior. FG’s behave similarly in every molecule they are part of. The chemistry of the organic molecule is defined by the function groups it contains 1 C C Alkanes Carbon - Carbon Multiple Bonds Carbon-heteroatom single bonds basic C N C C C X X= F, Cl, Br, I amines Alkenes Alkyl Halide H C C C O C C O Alkynes alcohols ethers acidic H H H C S C C C C S C C H sulfides C C thiols (disulfides) H H Arenes Carbonyl-oxygen double bonds (carbonyls) Carbon-nitrogen multiple bonds acidic basic O O O N H C H C O C Cl imine (Schiff base) aldehyde carboxylic acid acid chloride O O O O C C N C C C C O O C C nitrile (cyano group) ketones ester anhydrides O C N amide opsin Lys-NH2 + Lys- opsin H O H N rhodopsin H 2 Alkanes and Alkane Isomers Alkanes: organic compounds with only C-C and C-H single (s) bonds. general formula for alkanes: CnH(2n+2) Saturated hydrocarbons Hydrocarbons: contains only carbon and hydrogen Saturated" contains only single bonds Isomers: compounds with the same chemical formula, but different arrangement of atoms Constitutional isomer: have different connectivities (not limited to alkanes) C H O C4H10 C5H12 2 6 O OH butanol diethyl ether straight-chain or normal hydrocarbons branched hydrocarbons n-butane n-pentane Systematic Nomenclature (IUPAC System) Prefix-Parent-Suffix