Grade Dysplasia in Laser Capture Microdissected Archival Tissue
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Gene Symbol Category ACAN ECM ADAM10 ECM Remodeling-Related ADAM11 ECM Remodeling-Related ADAM12 ECM Remodeling-Related ADAM15 E
Supplementary Material (ESI) for Integrative Biology This journal is (c) The Royal Society of Chemistry 2010 Gene symbol Category ACAN ECM ADAM10 ECM remodeling-related ADAM11 ECM remodeling-related ADAM12 ECM remodeling-related ADAM15 ECM remodeling-related ADAM17 ECM remodeling-related ADAM18 ECM remodeling-related ADAM19 ECM remodeling-related ADAM2 ECM remodeling-related ADAM20 ECM remodeling-related ADAM21 ECM remodeling-related ADAM22 ECM remodeling-related ADAM23 ECM remodeling-related ADAM28 ECM remodeling-related ADAM29 ECM remodeling-related ADAM3 ECM remodeling-related ADAM30 ECM remodeling-related ADAM5 ECM remodeling-related ADAM7 ECM remodeling-related ADAM8 ECM remodeling-related ADAM9 ECM remodeling-related ADAMTS1 ECM remodeling-related ADAMTS10 ECM remodeling-related ADAMTS12 ECM remodeling-related ADAMTS13 ECM remodeling-related ADAMTS14 ECM remodeling-related ADAMTS15 ECM remodeling-related ADAMTS16 ECM remodeling-related ADAMTS17 ECM remodeling-related ADAMTS18 ECM remodeling-related ADAMTS19 ECM remodeling-related ADAMTS2 ECM remodeling-related ADAMTS20 ECM remodeling-related ADAMTS3 ECM remodeling-related ADAMTS4 ECM remodeling-related ADAMTS5 ECM remodeling-related ADAMTS6 ECM remodeling-related ADAMTS7 ECM remodeling-related ADAMTS8 ECM remodeling-related ADAMTS9 ECM remodeling-related ADAMTSL1 ECM remodeling-related ADAMTSL2 ECM remodeling-related ADAMTSL3 ECM remodeling-related ADAMTSL4 ECM remodeling-related ADAMTSL5 ECM remodeling-related AGRIN ECM ALCAM Cell-cell adhesion ANGPT1 Soluble factors and receptors -

Ultra-Processed Food Targets Bone Quality Via Endochondral Ossification
Bone Research www.nature.com/boneres ARTICLE OPEN Ultra-processed food targets bone quality via endochondral ossification Janna Zaretsky1, Shelley Griess-Fishheimer1, Adi Carmi1, Tamara Travinsky Shmul1, Lior Ofer2, Tali Sinai1, Svetlana Penn1, Ron Shahar2 and Efrat Monsonego-Ornan 1 Ultra-processed foods have known negative implications for health; however, their effect on skeletal development has never been explored. Here, we show that young rats fed ultra-processed food rich in fat and sugar suffer from growth retardation due to lesions in their tibial growth plates. The bone mineral density decreases significantly, and the structural parameters of the bone deteriorate, presenting a sieve-like appearance in the cortices and poor trabecular parameters in long bones and vertebrae. This results in inferior mechanical performance of the entire bone with a high fracture risk. RNA sequence analysis of the growth plates demonstrated an imbalance in extracellular matrix formation and degradation and impairment of proliferation, differentiation and mineralization processes. Our findings highlight, for the first time, the severe impact of consuming ultra-processed foods on the growing skeleton. This pathology extends far beyond that explained by the known metabolic effects, highlighting bone as a new target for studies of modern diets. Bone Research (2021) ;9:14 https://doi.org/10.1038/s41413-020-00127-9 1234567890();,: INTRODUCTION penetration of blood vessels and bone tissue formation by The vertebrate skeleton has evolved as a dynamic system that osteoblasts.1,2,4,6 The EO process is tightly orchestrated by various serves numerous functions, such as protecting internal organs, signaling molecules and transcription factors, including transform- creating attachment sites for muscles to produce locomotion, ing growth factor β/bone morphogenetic protein (BMP), fibroblast providing a reservoir for minerals, and serving as a hematopoietic growth factor (FGF), Wnt, hedgehog, and the transcription factor niche. -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. -

Identification, by Systematic RNA Sequencing, of Novel Candidate
Laboratory Investigation (2015) 95, 1077–1088 © 2015 USCAP, Inc All rights reserved 0023-6837/15 Identification, by systematic RNA sequencing, of novel candidate biomarkers and therapeutic targets in human soft tissue tumors Anne E Sarver1, Aaron L Sarver2, Venugopal Thayanithy1 and Subbaya Subramanian1 Human sarcomas comprise a heterogeneous group of more than 50 subtypes broadly classified into two groups: bone and soft tissue sarcomas. Such heterogeneity and their relative rarity have made them challenging targets for classification, biomarker identification, and development of improved treatment strategies. In this study, we used RNA sequencing to analyze 35 primary human tissue samples representing 13 different sarcoma subtypes, along with benign schwannoma, and normal bone and muscle tissues. For each sarcoma subtype, we detected unique messenger RNA (mRNA) expression signatures, which we further subjected to bioinformatic functional analysis, upstream regulatory analysis, and microRNA (miRNA) targeting analysis. We found that, for each sarcoma subtype, significantly upregulated genes and their deduced upstream regulators included not only previously implicated known players but also novel candidates not previously reported to be associated with sarcoma. For example, the schwannoma samples were characterized by high expression of not only the known associated proteins GFAP and GAP43 but also the novel player GJB6. Further, when we integrated our expression profiles with miRNA expression data from each sarcoma subtype, we were able to deduce potential key miRNA–gene regulator relationships for each. In the Ewing’s sarcoma and fibromatosis samples, two sarcomas where miR-182-5p is significantly downregulated, multiple predicted targets were significantly upregulated, including HMCN1, NKX2-2, SCNN1G, and SOX2. -

New and Paradoxical Roles of Matrix Metalloproteinases in the Tumor Microenvironment
MINI REVIEW ARTICLE published: 17 July 2012 doi: 10.3389/fphar.2012.00140 New and paradoxical roles of matrix metalloproteinases in the tumor microenvironment Agnès Noël 1*, Ana Gutiérrez-Fernández 2, Nor Eddine Sounni 1, Niels Behrendt 3,4, Erik Maquoi 1, Ida K. Lund 3,4, Santiago Cal 2, Gunilla Hoyer-Hansen3,4 and Carlos López-Otín2 1 Laboratory of Tumor and Development Biology, GIGA-Cancer, University of Liège, Liège, Belgium 2 Departamento de Bioquímica y Biología Molecular, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Asturias, Spain 3 The Finsen Laboratory, Copenhagen University Hospital, Copenhagen Biocenter, Copenhagen N, Denmark 4 Biotech Research & Innovation Centre, University of Copenhagen, Copenhagen Biocenter, Copenhagen N, Denmark Edited by: Processes such as cell proliferation, angiogenesis, apoptosis, or invasion are strongly influ- Pierre Sonveaux, University of enced by the surrounding microenvironment of the tumor. Therefore, the ability to change Louvain Medical School, Belgium these surroundings represents an important property through which tumor cells are able to Reviewed by: Juan Iovanna, INSERM, France acquire specific functions necessary for tumor growth and dissemination. Matrix metallo- Hervé Emonard, CNRS, France proteinases (MMPs) constitute key players in this process, allowing tumor cells to modify Cyril Rauch, Nottingham University, the extracellular matrix (ECM) and release cytokines, growth factors, and other cell-surface UK molecules, ultimately facilitating protease-dependent tumor progression. Remodeling of *Correspondence: the ECM by collagenolytic enzymes such as MMP1, MMP8, MMP13, or the membrane- Agnès Noël, University of Liège, Laboratory of Tumour & Development bound MT1-MMP as well as by other membrane-anchored proteases is required for invasion Biology, Tour de Pathologie, B23, Sart and recruitment of novel blood vessels. -

The Metalloproteinase-Proteoglycans ADAMTS7 and ADAMTS12 Provide an Innate, Tendon-Specific Protective Mechanism Against Heterotopic Ossification
RESEARCH ARTICLE The metalloproteinase-proteoglycans ADAMTS7 and ADAMTS12 provide an innate, tendon-specific protective mechanism against heterotopic ossification Timothy J. Mead,1 Daniel R. McCulloch,1 Jason C. Ho,1,2 Yaoyao Du,1 Sheila M. Adams,3 David E. Birk,3 and Suneel S. Apte1 1Department of Biomedical Engineering and the Orthopaedic and Rheumatologic Institute, Cleveland Clinic Lerner Research Institute, Cleveland, Ohio, USA. 2Department of Orthopaedic Surgery and the Orthopaedic and Rheumatology Institute, Cleveland Clinic, Cleveland, Ohio, USA. 3Departments of Molecular Pharmacology and Physiology and Orthopaedics and Sports Medicine, University of South Florida, Morsani College of Medicine, Tampa, Florida, USA. Heterotopic ossification (HO) is a significant clinical problem with incompletely resolved mechanisms. Here, the secreted metalloproteinases ADAMTS7 and ADAMTS12 are shown to comprise a unique proteoglycan class that protects against a tendency toward HO in mouse hindlimb tendons, menisci, and ligaments. Adamts7 and Adamts12 mRNAs were sparsely expressed in murine forelimbs but strongly coexpressed in hindlimb tendons, skeletal muscle, ligaments, and meniscal fibrocartilage. Adamts7–/– Adamts12–/– mice, but not corresponding single-gene mutants, which demonstrated compensatory upregulation of the intact homolog mRNA, developed progressive HO in these tissues after 4 months of age. Adamts7–/– Adamts12–/– tendons had abnormal collagen fibrils, accompanied by reduced levels of the small leucine-rich proteoglycans (SLRPs) biglycan, fibromodulin, and decorin, which regulate collagen fibrillogenesis. Bgn–/0 Fmod–/– mice are known to have a strikingly similar hindlimb HO to that of Adamts7–/– Adamts12–/– mice, implicating fibromodulin and biglycan reduction as a likely mechanism underlying HO in Adamts7–/– Adamts12–/– mice. Interestingly, degenerated human biceps tendons had reduced ADAMTS7 mRNA compared with healthy biceps tendons, which expressed both ADAMTS7 and ADAMTS12. -

ADAMTS12 Acts As a Tumor Microenvironment Related Cancer Promoter in Gastric Cancer Yangming Hou1, Yingjuan Xu2 & Dequan Wu1*
www.nature.com/scientificreports OPEN ADAMTS12 acts as a tumor microenvironment related cancer promoter in gastric cancer Yangming Hou1, Yingjuan Xu2 & Dequan Wu1* The infltration degree of immune and stromal cells has been shown clinically signifcant in tumor microenvironment (TME). However, the utility of stromal and immune components in Gastric cancer (GC) has not been investigated in detail. In the present study, ESTIMATE and CIBERSORT algorithms were applied to calculate the immune/stromal scores and the proportion of tumor-infltrating immune cell (TIC) in GC cohort, including 415 cases from The Cancer Genome Atlas (TCGA) database. The diferentially expressed genes (DEGs) were screened by Cox proportional hazard regression analysis and protein–protein interaction (PPI) network construction. Then ADAMTS12 was regarded as one of the most predictive factors. Further analysis showed that ADAMTS12 expression was signifcantly higher in tumor samples and correlated with poor prognosis. Gene Set Enrichment Analysis (GSEA) indicated that in high ADAMTS12 expression group gene sets were mainly enriched in cancer and immune-related activities. In the low ADAMTS12 expression group, the genes were enriched in the oxidative phosphorylation pathway. CIBERSORT analysis for the proportion of TICs revealed that ADAMTS12 expression was positively correlated with Macrophages M0/M1/M2 and negatively correlated with T cells follicular helper. Therefore, ADAMTS12 might be a tumor promoter and responsible for TME status and tumor energy metabolic conversion. According to the latest global cancer epidemic statistics (GLOBOCAN), gastric cancer (GC) is the third lead- ing cause of cancer related death worldwide1,2. GC patients are frequently diagnosed at advanced stage with the fve-year survival rate less than 20%3. -

SUPPLEMENTARY TABLE 3. Gene Ranking: 277 Deregulated Genes
SUPPLEMENTARY TABLE 3. Gene ranking: 277 deregulated genes. overexpressed in the sequence NCF/CAF-PT/CAF-LM mean Log2 normalised expression Fold changes Probe ID gene symbol NCF CAF-PT CAF-LM CAF-PT/NCF CAF-LM/CAF-PT pval qval Gene_description 8114970 C5orf46 4.721 5.843 9.288 1.773 14.845 4.22E-05 0 chromosome 5 open reading frame 46 8130173 RAET1L 7.421 8.139 8.923 1.667 1.665 4.22E-05 0 retinoic acid early transcript 1L 7898413 PADI1 6.316 6.989 7.628 1.535 1.590 4.22E-05 0 peptidyl arginine deiminase, type I 8020455 GATA6 7.902 8.665 9.846 1.598 2.751 4.22E-05 0 GATA binding protein 6 8058450 GPR1 5.420 6.989 8.559 2.921 3.498 4.22E-05 0 G protein-coupled receptor 1 8111387 ADAMTS12 8.507 9.214 10.544 1.663 2.451 4.22E-05 0 ADAM metallopeptidase with thrombospondin type 1 motif, 12 8100382 CHIC2 8.343 9.081 10.723 1.616 3.392 4.22E-05 0 cysteine-rich hydrophobic domain 2 7938485 MICAL2 9.309 10.477 11.568 2.320 2.180 4.22E-05 0 microtubule associated monoxygenase, calponin and LIM domain containing 2 7909789 TGFB2 8.478 10.294 11.406 4.254 1.710 4.22E-05 0 transforming growth factor, beta 2 7953284 NTF3 7.510 8.920 9.474 2.429 1.601 4.22E-05 0 neurotrophin 3 8110932 SEMA5A 8.892 10.081 11.490 2.956 2.526 4.22E-05 0 semaphorin 5A 8122720 ULBP2 6.024 7.831 8.695 3.896 1.531 4.22E-05 0 UL16 binding protein 2 7945245 NTM 7.702 9.005 10.360 1.976 2.891 4.22E-05 0 neurotrimin 8020878 MAPRE2 9.217 9.644 11.028 1.508 2.648 4.22E-05 0 microtubule-associated protein, RP/EB family, member 2 8112007 EMB 4.721 5.986 7.610 1.941 4.055 4.22E-05 0 -

Supplementary Figures
Supplementary Figures Fig. S1. Distribution of total mutation pool. a) Statistics of different mutational types. b) The distribution of mutational allele frequency. c) Circle graph illustrating the chromosomal assignment of total mutation pool. The outer ring provides an overview of detected ctDNA mutations in each chromosome, while the inner ring shows the mutation burden of each chromosome. 1 Fig. S2. DNMT3A mutations in the experimental cohort. The upper graph shows all 19 DNMT3A mutations and their protein localization. The red rectangle indicates DNA_methylase domains. The lollipop graph is generated by online tools of cbioportal (http://www.cbioportal.org/visualize). The lower graph illustrates the diagnostic age distribution for patients with and without DNMT3A mutations. The p value is calculated by nonparametric t-test. 2 Fig. S3. Overview of clinical actionability for patients with drug-sensitive and resistant mutations. Each horizontal column represents an individual patients. Columns with different colors indicate diverse actionable mutations, and the length of column represents the number of mutations. Patients are divided into three groups using horizontal dotted lines, according to the mutation types. 3 Fig. S4. Concordance and mutant allele frequencies for matched sample pairs. Venn diagrams present the mutation overlap between matched tissue and blood samples. Scatter diagrams show the correlation between the allele frequencies of overlapping mutations in ctDNA vs. tissue DNA. Each dot represents an overlapping mutation, -

Transcriptome Profiling Reveals the Complexity of Pirfenidone Effects in IPF
ERJ Express. Published on August 30, 2018 as doi: 10.1183/13993003.00564-2018 Early View Original article Transcriptome profiling reveals the complexity of pirfenidone effects in IPF Grazyna Kwapiszewska, Anna Gungl, Jochen Wilhelm, Leigh M. Marsh, Helene Thekkekara Puthenparampil, Katharina Sinn, Miroslava Didiasova, Walter Klepetko, Djuro Kosanovic, Ralph T. Schermuly, Lukasz Wujak, Benjamin Weiss, Liliana Schaefer, Marc Schneider, Michael Kreuter, Andrea Olschewski, Werner Seeger, Horst Olschewski, Malgorzata Wygrecka Please cite this article as: Kwapiszewska G, Gungl A, Wilhelm J, et al. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF. Eur Respir J 2018; in press (https://doi.org/10.1183/13993003.00564-2018). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2018 Copyright 2018 by the European Respiratory Society. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF Grazyna Kwapiszewska1,2, Anna Gungl2, Jochen Wilhelm3†, Leigh M. Marsh1, Helene Thekkekara Puthenparampil1, Katharina Sinn4, Miroslava Didiasova5, Walter Klepetko4, Djuro Kosanovic3, Ralph T. Schermuly3†, Lukasz Wujak5, Benjamin Weiss6, Liliana Schaefer7, Marc Schneider8†, Michael Kreuter8†, Andrea Olschewski1, -
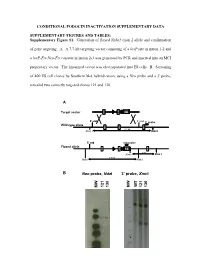
Supplementary Figure S1. Generation of Floxed Nphs2 Exon 2 Allele and Confirmation
CONDITIONAL PODOCIN INACTIVATION SUPPLEMENTARY DATA SUPPLEMENTARY FIGURES AND TABLES: Supplementary Figure S1. Generation of floxed Nphs2 exon 2 allele and confirmation of gene targeting. A. A 7.7-kb targeting vector consisting of a loxP site in intron 1-2 and a loxP-Frt-Neo-Frt cassette in intron 2-3 was generated by PCR and inserted into an MCI proprietary vector. The linearized vector was electroporated into ES cells. B. Screening of 400 ES cell clones by Southern blot hybridization, using a Neo probe and a 3' probe, revealed two correctly targeted clones 121 and 130. A Target vector 1 2 NEO 5’ end 3’ end 3’ probe Wild-type allele 1 2 3 9.2 kb Xmn I Xmn I 5’ end neo probe Floxed allele 1 2 NEO 3 5 kb Xmn I Xmn I 7.8 kb Nde I Nde I B Neo probe, NdeI 3’ probe, XmnI WT 121 130 121 130 MW MW - 1 - Supplementary Figure S2. Generation of Nphs2lox2/-,Cre+ mice and excision of exon 2 upon Cre recombinase induction. A. Triallelic Nphs2lox2/-,Cre+ mice were obtained by mating phenotypically normal Nphs2lox2/lox2 mice with Nphs2+/-,Cre+ mice. Mendelian inheritance of these alleles was observed. B. Genotypes were verified by multiplex PCR of tail genomic DNA. C. Cre recombinase activity was induced upon tamoxifen administration, leading to excision of the floxed exon 2 of the Nphs2 gene. D. Cre recombinase activity in the kidney was verified by PCR using a set of forward and reverse primers designed around exon 2 and demonstrating a 692-bp product (before Cre) and a 316-bp product (after Cre) using genomic DNA extracted from the renal cortex.