Bacterial Classification, Structure and Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 10: Classification of Microorganisms
Chapter 10: Classification of Microorganisms 1. The Taxonomic Hierarchy 2. Methods of Identification 1. The Taxonomic Hierarchy Phylogenetic Tree of the 3 Domains Taxonomic Hierarchy • 8 successive taxa are used to classify each species: Domain Kingdom Phylum Class Order Family Genus **species can also contain different strains** Species Scientific Nomenclature To avoid confusion, every type of organism must be referred to in a consistent way. The current system of nomenclature (naming) has been in use since the 18th century: • every type of organism is referred by its genus name followed by its specific epithet (i.e., species name) Homo sapiens (H. sapiens) Escherichia coli (E. coli) • name should be in italics and only the genus is capitalized which can also be abbreviated • names are Latin (or “Latinized” Greek) with the genus being a noun and the specific epithet an adjective **strain info can be listed after the specific epithet (e.g., E. coli DH5α)** 2. Methods of Identification Biochemical Testing In addition to morphological (i.e., appearance under the microscope) and differential staining characteristics, microorganisms can also be identified by their biochemical “signatures”: • the nutrient requirements and metabolic “by-products” of of a particular microorganism • different growth media can be used to test the physiological characteristics of a microorganism • e.g., medium with lactose only as energy source • e.g., medium that reveals H2S production **appearance on test medium reveals + or – result!** Commercial devices for rapid Identification Perform multiple tests simultaneously Enterotube II Such devices involve the simultaneous inoculation of various test media: • ~24 hrs later the panel of results reveals ID of organism! Use of Dichotomous Keys Series of “yes/no” biochemical tests to ID organism. -

The Syntrophy Hypothesis for the Origin of Eukaryotes Revisited Purificación López-García, David Moreira
The Syntrophy hypothesis for the origin of eukaryotes revisited Purificación López-García, David Moreira To cite this version: Purificación López-García, David Moreira. The Syntrophy hypothesis for the origin of eukaryotes revisited. Nature Microbiology, Nature Publishing Group, 2020, 5 (5), pp.655-667. 10.1038/s41564- 020-0710-4. hal-02988531 HAL Id: hal-02988531 https://hal.archives-ouvertes.fr/hal-02988531 Submitted on 3 Dec 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 2 Perspectives 3 4 5 6 The Syntrophy hypothesis for the origin of eukaryotes revisited 7 8 Purificación López-García1 and David Moreira1 9 10 1 Ecologie Systématique Evolution, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France 11 12 13 14 *Correspondence to: [email protected] 15 16 17 18 19 1 20 The discovery of Asgard archaea, phylogenetically closer to eukaryotes than other archaea, together with 21 improved knowledge of microbial ecology impose new constraints on emerging models for the origin of the 22 eukaryotic cell (eukaryogenesis). Long-held views are metamorphosing in favor of symbiogenetic models 23 based on metabolic interactions between archaea and bacteria. These include the classical Searcy’s and 24 hydrogen hypothesis, and the more recent Reverse Flow and Entangle-Engulf-Enslave (E3) models. -

Intelligent Design, Abiogenesis, and Learning from History: Dennis R
Author Exchange Intelligent Design, Abiogenesis, and Learning from History: Dennis R. Venema A Reply to Meyer Dennis R. Venema Weizsäcker’s book The World View of Physics is still keeping me very busy. It has again brought home to me quite clearly how wrong it is to use God as a stop-gap for the incompleteness of our knowledge. If in fact the frontiers of knowledge are being pushed back (and that is bound to be the case), then God is being pushed back with them, and is therefore continually in retreat. We are to find God in what we know, not in what we don’t know; God wants us to realize his presence, not in unsolved problems but in those that are solved. Dietrich Bonhoeffer1 am thankful for this opportunity to nature, is the result of intelligence. More- reply to Stephen Meyer’s criticisms over, this assertion is proffered as the I 2 of my review of his book Signature logical basis for inferring design for the in the Cell (hereafter Signature). Meyer’s origin of biological information: if infor- critiques of my review fall into two gen- mation only ever arises from intelli- eral categories. First, he claims I mistook gence, then the mere presence of Signature for an argument against bio- information demonstrates design. A few logical evolution, rendering several of examples from Signature make the point my arguments superfluous. Secondly, easily: Meyer asserts that I have failed to refute … historical scientists can show that his thesis by not providing a “causally a presently acting cause must have adequate alternative explanation” for the been present in the past because the origin of life in that the few relevant cri- proposed candidate is the only known tiques I do provide are “deeply flawed.” cause of the effect in question. -

Revised Glossary for AQA GCSE Biology Student Book
Biology Glossary amino acids small molecules from which proteins are A built abiotic factor physical or non-living conditions amylase a digestive enzyme (carbohydrase) that that affect the distribution of a population in an breaks down starch ecosystem, such as light, temperature, soil pH anaerobic respiration respiration without using absorption the process by which soluble products oxygen of digestion move into the blood from the small intestine antibacterial chemicals chemicals produced by plants as a defence mechanism; the amount abstinence method of contraception whereby the produced will increase if the plant is under attack couple refrains from intercourse, particularly when an egg might be in the oviduct antibiotic e.g. penicillin; medicines that work inside the body to kill bacterial pathogens accommodation ability of the eyes to change focus antibody protein normally present in the body acid rain rain water which is made more acidic by or produced in response to an antigen, which it pollutant gases neutralises, thus producing an immune response active site the place on an enzyme where the antimicrobial resistance (AMR) an increasing substrate molecule binds problem in the twenty-first century whereby active transport in active transport, cells use energy bacteria have evolved to develop resistance against to transport substances through cell membranes antibiotics due to their overuse against a concentration gradient antiretroviral drugs drugs used to treat HIV adaptation features that organisms have to help infections; they -

Marine Microorganisms: Evolution and Solution to Pollution Fu L Li1, Wang B1,2
COMMENTARY Marine microorganisms: Evolution and solution to pollution Fu L Li1, Wang B1,2 Li FL, Wang B. Marine microorganisms: Evolution and solution to pollution. J Mar Microbiol. 2018;2(1):4-5. nce ocean nurtured life, now she needs our care. Marine microorganism will be an opportunity to further understand ourselves and to seek for new Ois the host of ocean in all ages. We should learn from them humbly. methods of fighting old infections. Marine microorganism is tightly bond with human during the history of evolution and nowadays’ environment pollution. Along with industrial revolution, our marine ecosystem suffered serious pollutions. Microplastics are tiny plastic particles (<5 mm) (Figure 1B), Although the topic is still in debate, life is probably originated from which poison marine lives. Because these microplastics are very hard to be submarine in hydrothermal vent systems (1). In the journey of evolution, our degraded, it is predicted that there will be more microplastics than fish in biosphere was completely dominated by microbes for a very long time (Figure ocean by the year 2050 (7). Since marine sediments are considered as the sink 1A). Human being evolves with those microorganisms. Consequently, of microplastics and marine microbes are key dwellers of marine sediments, the influences of microorganisms can be found in all aspects of human more attention should be paid on the interactions between microplastics biology. More than 65% of our genes originated with bacteria, archaea, and and marine microbes. Actually, a call for this has been published in 2011 unicellular eukaryotes, including those genes responsible for host-microbe (8). -

The Hidden Kingdom
INTRODUCTION Fungi—The Hidden Kingdom OBJECTIVE • To provide students with basic knowledge about fungi Activity 0.1 BACKGROUND INFORMATION The following text provides an introduction to the fungi. It is written with the intention of sparking curiosity about this GRADES fascinating biological kingdom. 4-6 with a K-3 adaptation TEACHER INSTRUCTIONS TYPE OF ACTIVITY 1. With your class, brainstorm everything you know about fungi. Teacher read/comprehension 2. For younger students, hand out the question sheet before you begin the teacher read and have them follow along and MATERIALS answer the questions as you read. • copies of page 11 3. For older students, inform them that they will be given a • pencils brainteaser quiz (that is not for evaluation) after you finish reading the text. VOCABULARY 4. The class can work on the questions with partners or in groups bioremediation and then go over the answers as a class. Discuss any chitin particularly interesting facts and encourage further fungi independent research. habitat hyphae K-3 ADAPTATION kingdom 1. To introduce younger students to fungi, you can make a KWL lichens chart either as a class or individually. A KWL chart is divided moulds into three parts. The first tells what a student KNOWS (K) mushrooms about a subject before it is studied in class. The second part mycelium tells what the student WANTS (W) to know about that subject. mycorrhizas The third part tells what the child LEARNED (L) after studying nematodes that subject. parasitic fungi 2. Share some of the fascinating fungal facts presented in the photosynthesis “Fungi—The Hidden Kingdom” text with your students. -

Framing Major Prebiotic Transitions As Stages of Protocell Development: Three Challenges for Origins-Of-Life Research
Framing major prebiotic transitions as stages of protocell development: three challenges for origins-of-life research Ben Shirt-Ediss1, Sara Murillo-Sánchez2,3 and Kepa Ruiz-Mirazo*2,3 Commentary Open Access Address: Beilstein J. Org. Chem. 2017, 13, 1388–1395. 1Interdisciplinary Computing and Complex BioSystems Group, doi:10.3762/bjoc.13.135 University of Newcastle, UK, 2Dept. Logic and Philosophy of Science, University of the Basque Country, Spain and 3Biofisika Institute Received: 16 February 2017 (CSIC, UPV-EHU), Spain Accepted: 27 June 2017 Published: 13 July 2017 Email: Kepa Ruiz-Mirazo* - [email protected] This article is part of the Thematic Series "From prebiotic chemistry to molecular evolution". * Corresponding author Guest Editor: L. Cronin Keywords: functional integration; origins of life; prebiotic evolution; protocells © 2017 Shirt-Ediss et al.; licensee Beilstein-Institut. License and terms: see end of document. Abstract Conceiving the process of biogenesis as the evolutionary development of highly dynamic and integrated protocell populations provides the most appropriate framework to address the difficult problem of how prebiotic chemistry bridged the gap to full-fledged living organisms on the early Earth. In this contribution we briefly discuss the implications of taking dynamic, functionally inte- grated protocell systems (rather than complex reaction networks in bulk solution, sets of artificially evolvable replicating molecules, or even these same replicating molecules encapsulated in passive compartments) -

Advanced Morphometric Techniques Applied to The
UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIEROS DE TELECOMUNICACIÓN ADVANCED MORPHOMETRIC TECHNIQUES APPLIED TO THE STUDY OF HUMAN BRAIN ANATOMY TESIS DOCTORAL Yasser Alemán Gómez Ingeniero en Tecnologías Nucleares y Energéticas Máster en Neurociencias Madrid, 2015 DEPARTAMENTO DE INGENIERÍA ELECTRÓNICA ESCUELA TÉCNICA SUPERIOR DE INGENIEROS DE TELECOMUNICACIÓN PHD THESIS ADVANCED MORPHOMETRIC TECHNIQUES APPLIED TO THE STUDY OF HUMAN BRAIN ANATOMY AUTHOR Yasser Alemán Gómez Ing. en Tecnologías Nucleares y Energéticas MSc en Neurociencias ADVISOR Manuel Desco Menéndez, MScE, MD, PhD Madrid, 2015 Departamento de Ingeniería Electrónica Escuela Técnica Superior de Ingenieros de Telecomunicación Universidad Politécnica de Madrid Ph.D. Thesis Advanced morphometric techniques applied to the study of human brain anatomy Tesis doctoral Técnicas avanzadas de morfometría aplicadas al estudio de la anatomía cerebral humana Author: Yasser Alemán Gómez Advisor: Manuel Desco Menéndez Committee: Andrés Santos Lleó Universidad Politécnica de Madrid, Madrid, Spain Javier Pascau Gonzalez-Garzón Universidad Carlos III de Madrid, Madrid, Spain Raymond Salvador Civil FIDMAG – Germanes Hospitalàries, Barcelona, Spain Pablo Campo Martínez-Lage Universidad Autónoma de Madrid, Madrid, Spain Juan Domingo Gispert López Universidad Pompeu Fabra, Barcelona, Spain María Jesús Ledesma Carbayo Universidad Politécnica de Madrid, Madrid, Spain Juan José Vaquero López Universidad Carlos III de Madrid, Madrid, Spain Esta Tesis ha sido desarrollada en el Laboratorio de Imagen Médica de la Unidad de Medicina y Cirugía Experimental del Instituto de Investigación Sanitaria Gregorio Marañón y en colaboración con el Servicio de Psiquiatría del Niño y del Adolescente del Departamento de Psiquiatría del Hospital General Universitario Gregorio Marañón de Madrid, España. Tribunal nombrado por el Sr. -

Introduction to Bacteriology and Bacterial Structure/Function
INTRODUCTION TO BACTERIOLOGY AND BACTERIAL STRUCTURE/FUNCTION LEARNING OBJECTIVES To describe historical landmarks of medical microbiology To describe Koch’s Postulates To describe the characteristic structures and chemical nature of cellular constituents that distinguish eukaryotic and prokaryotic cells To describe chemical, structural, and functional components of the bacterial cytoplasmic and outer membranes, cell wall and surface appendages To name the general structures, and polymers that make up bacterial cell walls To explain the differences between gram negative and gram positive cells To describe the chemical composition, function and serological classification as H antigen of bacterial flagella and how they differ from flagella of eucaryotic cells To describe the chemical composition and function of pili To explain the unique chemical composition of bacterial spores To list medically relevant bacteria that form spores To explain the function of spores in terms of chemical and heat resistance To describe characteristics of different types of membrane transport To describe the exact cellular location and serological classification as O antigen of Lipopolysaccharide (LPS) To explain how the structure of LPS confers antigenic specificity and toxicity To describe the exact cellular location of Lipid A To explain the term endotoxin in terms of its chemical composition and location in bacterial cells INTRODUCTION TO BACTERIOLOGY 1. Two main threads in the history of bacteriology: 1) the natural history of bacteria and 2) the contagious nature of infectious diseases, were united in the latter half of the 19th century. During that period many of the bacteria that cause human disease were identified and characterized. 2. Individual bacteria were first observed microscopically by Antony van Leeuwenhoek at the end of the 17th century. -
Lokiarchaeota: Biologists Discover 'Missing Link' Microorganism
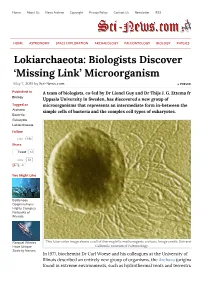
Home About Us News Archive Copyright Privacy Policy Contact Us Newsletter RSS HOME ASTRONOMY SPACE EXPLORATION ARCHAEOLOGY PALEONTOLOGY BIOLOGY PHYSICS Lokiarchaeota: Biologists Discover ‘Missing Link’ Microorganism May 7, 2015 by Sci-News.com « PREVIOUS Published in A team of biologists, co-led by Dr Lionel Guy and Dr Thijs J. G. Ettema from Biology Uppsala University in Sweden, has discovered a new group of Tagged as microorganisms that represents an intermediate form in-between the Archaea simple cells of bacteria and the complex cell types of eukaryotes. Bacteria Eukaryote Lokiarchaeota Follow Like 16k Share Tweet 12 Like 58 41 You Might Like Bottlenose Dolphins Form Highly Complex Networks of Friends Rorqual Whales This false-color image shows a cell of thermophilic methanogenic archaea. Image credit: University of Have Unique California Museum of Paleontology. Stretchy Nerves In 1977, biochemist Dr Carl Woese and his colleagues at the University of Illinois described an entirely new group of organisms, the Archaea (originally found in extreme environments, such as hydrothermal vents and terrestrial hot springs). The scientists were studying relationships among the prokaryotes using DNA Extinction of sequences, and found that Archaea have distinct molecular characteristics World’s Largest separating them from bacteria as well as from eukaryotes. They proposed that Herbivores May Lead to Empty life can be divided into three domains: Eukaryota, Eubacteria, and Landscapes, Say Archaebacteria. Researchers Despite that archaeal cells were simple and small like bacteria, scientists found that Archaea were more closely related to organisms with complex cell types, a group collectively known as ‘eukaryotes.’ This observation has puzzled Sichuan Bush biologists for years. -

A Practical Introduction to Landmark-Based Geometric Morphometrics
A PRACTICAL INTRODUCTION TO LANDMARK-BASED GEOMETRIC MORPHOMETRICS MARK WEBSTER Department of the Geophysical Sciences, University of Chicago, 5734 South Ellis Avenue, Chicago, IL 60637 and H. DAVID SHEETS Department of Physics, Canisius College, 2001 Main Street, Buffalo, NY 14208 ABSTRACT.—Landmark-based geometric morphometrics is a powerful approach to quantifying biological shape, shape variation, and covariation of shape with other biotic or abiotic variables or factors. The resulting graphical representations of shape differences are visually appealing and intuitive. This paper serves as an introduction to common exploratory and confirmatory techniques in landmark-based geometric morphometrics. The issues most frequently faced by (paleo)biologists conducting studies of comparative morphology are covered. Acquisition of landmark and semilandmark data is discussed. There are several methods for superimposing landmark configu- rations, differing in how and in the degree to which among-configuration differences in location, scale, and size are removed. Partial Procrustes superimposition is the most widely used superimposition method and forms the basis for many subsequent operations in geometric morphometrics. Shape variation among superimposed con- figurations can be visualized as a scatter plot of landmark coordinates, as vectors of landmark displacement, as a thin-plate spline deformation grid, or through a principal components analysis of landmark coordinates or warp scores. The amount of difference in shape between two configurations can be quantified as the partial Procrustes distance; and shape variation within a sample can be quantified as the average partial Procrustes distance from the sample mean. Statistical testing of difference in mean shape between samples using warp scores as variables can be achieved through a standard Hotelling’s T2 test, MANOVA, or canonical variates analysis (CVA). -

The Endosymbiotic Theory
Origin of Life The Endosymbiotic Theory Cleodie Swire The King's School, Canterbury, E-mail: [email protected] DOI: 10.4103/0974-6102.92200 Definitions Carbon dioxide + Water (with sunlight and chlorophyll) → Carbohydrate + Oxygen Prokaryote – Organism with cells without a true nucleus or other membrane-bound organelles Figure 2 shows that mitochondria and chloroplasts are very similar to prokaryotic cells; these observations Eukaryote – Organism whose cell(s) contain(s) a lead to The Endosymbiotic Theory. distinct, membrane-bound nucleus Theory Autotroph – An organism that can make its own food Researchers comparing the structures of prokaryotes Heterotroph – An organism that must obtain ready- and cell organelles, as shown in Figure 2, came to made food the conclusion that organelles such as mitochondria and chloroplasts had originally been bacteria that Endocytosis – A process in which a cell takes in were taken into larger bacteria by endocytosis and not materials by engulfing them and fusing them with its digested. The cells would have had a mutually beneficial membrane, as shown in Figure 1 (symbiotic) relationship. The ingested cells developed Aerobic – Organism that requires oxygen for survival Anaerobic – Organism that can function without oxygen Symbiosis – Two different organisms benefit from living and working together Endosymbiosis – One organism lives inside another Mitochondrion – Organelle where aerobic respiration occurs within the cell Carbohydrate + Oxygen → Carbon dioxide + Water + Energy Chloroplast – Organelle