BMC Genetics Biomed Central
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

View Entire Book
ODISHA REVIEW VOL. LXXI NO. 5 DECEMBER - 2014 MADHUSUDAN PADHI, I.A.S. Commissioner-cum-Secretary RANJIT KUMAR MOHANTY, O.A.S, ( SAG) Director DR. LENIN MOHANTY Editor Editorial Assistance Production Assistance Bibhu Chandra Mishra Debasis Pattnaik Bikram Maharana Sadhana Mishra Cover Design & Illustration D.T.P. & Design Manas Ranjan Nayak Hemanta Kumar Sahoo Photo Raju Singh Manoranjan Mohanty The Odisha Review aims at disseminating knowledge and information concerning Odisha’s socio-economic development, art and culture. Views, records, statistics and information published in the Odisha Review are not necessarily those of the Government of Odisha. Published by Information & Public Relations Department, Government of Odisha, Bhubaneswar - 751001 and Printed at Odisha Government Press, Cuttack - 753010. For subscription and trade inquiry, please contact : Manager, Publications, Information & Public Relations Department, Loksampark Bhawan, Bhubaneswar - 751001. Five Rupees / Copy E-mail : [email protected] Visit : http://odisha.gov.in Contact : 9937057528(M) CONTENTS Shrikshetra, Matha and its Impact Subhashree Mishra ... 1 Good Governance ... 3 India International Trade Fair - 2014 : An Overview Smita Kar ... 7 Mo Kahani' - The Memoir of Kunja Behari Dash : A Portrait Gallery of Pre-modern Rural Odisha Dr. Shruti Das ... 10 Protection of Fragile Ozone Layer of Earth Dr. Manas Ranjan Senapati ... 17 Child Labour : A Social Evil Dr. Bijoylaxmi Das ... 19 Reflections on Mahatma Gandhi's Life and Vision Dr. Brahmananda Satapathy ... 24 Christmas in Eternal Solitude Sonril Mohanty ... 27 Dr. B.R. Ambedkar : The Messiah of Downtrodden Rabindra Kumar Behuria ... 28 Untouchable - An Antediluvian Aspersion on Indian Social Stratification Dr. Narayan Panda ... 31 Kalinga, Kalinga and Kalinga Bijoyini Mohanty .. -

WP7 Biswamoy Pati
CORE Metadata, citation and similar papers at core.ac.uk Provided by LSE Research Online Asia Research Centre Working Paper 7 IDENTITY, HEGEMONY, RESISTANCE: CONVERSIONS IN ORISSA, 1800-2000 Dr Biswamoy Pati 1 Identity, Hegemony, Resistance: Conversions in Orissa œ 1800-2000 IDENTITY, HEGEMONY, RESISTANCE: CONVERSIONS IN ORISSA, 1800-2000* The Problem: It is widely held that as far as Hinduism is considered, the concept of conversion does not exist. Traditionally, it is said that one cannot become a Hindu by conversion, since one has to be born a Hindu and indeed, innumerable tracts, articles and books have been written on the subject. 1 However, as this paper will attempt to show, the ground realities of social dynamics would appear to suggest something different and this premise (that one cannot become a Hindu) may, in a profound sense, be open to contest. One can cite here the history of the adivasi peoples and their absorption into brahminical Hinduism - a social process that would, in fact, point to the gaps and contradictions existing between this kind of traditional belief (in the impossibility of ”conversion‘ to Hinduism) and the complexities of actual cultural practice on the ground. In other words, this paper seeks to argue that the history of the indigenous peoples would instead seem to suggest that, in an important sense, Hinduism did ”convert‘, and the paper further goes on to raise the question whether adivasis and outcastes were/are Hindus in the first place. 2 ”Common-sense‘ dictates that this absence of a system of conversion within Hinduism makes it by implication, more humane, tolerant and perhaps superior to proselytising religions like Islam and Christianity. -

Y-Chromosomal and Mitochondrial SNP Haplogroup Distribution In
Open Access Austin Journal of Forensic Science and Criminology Review Article Y-Chromosomal and Mitochondrial SNP Haplogroup Distribution in Indian Populations and its Significance in Disaster Victim Identification (DVI) - A Review Based Molecular Approach Sinha M1*, Rao IA1 and Mitra M2 1Department of Forensic Science, Guru Ghasidas Abstract University, India Disaster Victim Identification is an important aspect in mass disaster cases. 2School of Studies in Anthropology, Pt. Ravishankar In India, the scenario of disaster victim identification is very challenging unlike Shukla University, India any other developing countries due to lack of any organized government firm who *Corresponding author: Sinha M, Department of can make these challenging aspects an easier way to deal with. The objective Forensic Science, Guru Ghasidas University, India of this article is to bring spotlight on the potential and utility of uniparental DNA haplogroup databases in Disaster Victim Identification. Therefore, in this article Received: December 08, 2016; Accepted: January 19, we reviewed and presented the molecular studies on mitochondrial and Y- 2017; Published: January 24, 2017 chromosomal DNA haplogroup distribution in various ethnic populations from all over India that can be useful in framing a uniparental DNA haplogroup database on Indian population for Disaster Victim Identification (DVI). Keywords: Disaster Victim identification; Uniparental DNA; Haplogroup database; India Introduction with the necessity mentioned above which can reveal the fact that the human genome variation is not uniform. This inconsequential Disaster Victim Identification (DVI) is the recognized practice assertion put forward characteristics of a number of markers ranging whereby numerous individuals who have died as a result of a particular from its distribution in the genome, their power of discrimination event have their identity established through the use of scientifically and population restriction, to the sturdiness nature of markers to established procedures and methods [1]. -

Paika Rebellion : Heterogeneities and Linkages
International Journal of Asian Studies (2020), 17,1–18 doi:10.1017/S1479591420000157 ARTICLE “ ” . Genealogies of the Paika Rebellion : Heterogeneities and Linkages Akio Tanabe* The University of Tokyo *Corresponding author. Email: [email protected] Abstract The “Paika Rebellion” of 1817 in Orissa, India has been depicted by colonial officers as a local disturbance https://www.cambridge.org/core/terms caused by the dissatisfaction of one powerful individual deprived of traditional privileges who instigated the pāikas. The nationalist reconstruction has depicted the event as a popular freedom movement involv- ing various castes and classes of Orissan society. This has culminated in a current move to declare the “Paika Rebellion” the First Indian War of Independence. I would like to suggest a third perspective, which focuses on the heterogeneities and linkages of the Rebellion. It is important to note that the “Paika Rebellion” was a meeting point of plural genealogies: “tribal” revolts to protect autonomy, “peas- ant” resistance to secure livelihood, restorative attempts by the traditional landed class, and ruling class efforts to defend and expand authority. Appreciating the plural genealogies of the Rebellion leads to more perceptive understandings of the heterogeneous characteristics of popular movements and their aftermaths in modern India. Lastly, in order to go beyond colonial and dominant-caste centred perspec- tives, I propose that we name it the “Orissa Uprising of 1817”. Keywords: Paika Rebellion; India; Orissa; tribe; caste; nationalism; the First War of Independence , subject to the Cambridge Core terms of use, available at Introduction The year 2017 marked the bicentennial of what is called the “Paika Rebellion” that took place in coastal Orissa1 in 1817. -

Factional Politics in Orissa Since 1975
FACTIONAL POLITICS IN ORISSA SINCE 1975 Dissertation Submitted for tt)e Degree of Maittt of $Iitlo£(opl^p IN POLITICAL SCIENCE BY IFTEEHAR AHEMMED Under th« supervision of Prof. A. F. USMANI DEPARTMENT OF POLITICAL SCIENCE ALIGARH MUSLIM UNIVERSITY ALIGARH (INDIA) 1989 ( Office : 6720 Phone : ) Res. : 4342 A. F. USMANI f Office : 266 DEPARTMENT OF POLITICAL SCIENCE M.A., LL.B., Ph.D. ALIGARH MUSLIM UNIVERSITY, ALIGARH 202002, INDIA Professor & Chairman Octob^ 21, 19S9 Tka -i& io tOAtliy that tAn. I^tzkhoA Aheimed hcu, compl^tzd koi (LU,i,QjvtatLon zntotlzd: "FacXA^mZ Potituu In QUAA^O. ^Jjica 1975" andzn. my 6upeAv-u^on. The. wofik -Li onA.QlnaJi. I ($ce£ that the. d-u^e^tat^on i^ iuAtable (^oA ^abmi&^ton {jOn. the, de.gfiee Oj$ ^^cL&teA o^ ?hAZo6ophjj -in Votitlcal ScA.e.nce. ^rdZM^ ( A.F. USMAWI COflTENTS Page Acknowledg^neat Preface CHAPTER - I Introduction: Orissai The State and : 1-20 Its People CHAPTER - II Factionalism: Hypothetical Frame work : 21 - 33 CHAPTER - III Factionalism: A Historical Perspective: 34 - 62 CHAPTER - IV Mid-term Election of 1974 And Faction-: 63 - 87 alism in Orissa CHAPTER - V Factionalism During Janata Rule : 88 - 107 CHAPTER - VI Return to Congress System: The Era of :108 - 151 J.B, Patnaik. Conclusion :152 - 163 Bibliography :164 - 173 I mj>h to KZ-dofid my dzzp ^zmz o^ gAxitltadz to VKoiu/^ofi A.V. OAmcufu,, ChjaAJum.n,V2.p<Uitmtnt OjJ VotiticaZ Science, AtigaAh MiuLim UyuveAA^y, AtigaAh, Mho \s&n.y kAjtidly giUdzd me. In tkz pH.0QKe^& o{j thii viOKk. -

Council of Ministers of Odisha (1937–2020)
1 COUNCIL OF MINISTERS OF ODISHA (1937–2020) ODISHA LEGISLATIVE ASSEMBLY, BHUBANESWAR PRINTED AT ODISHA GOVERNMENT PRESS, MADHUPATNA, CUTTACK-10 COUNCIL OF MINISTERS OF ODISHA (1937–2020) ODISHA LEGISLATIVE ASSEMBLY BHUBANESWAR FIRST PRE-INDEPENDENT ASSEMBLY, 1937 Hon'ble Governor His Excellency Sir John Austen Hubback Hon'ble Speaker Shri Mukunda Prasad Das Hon'ble Deputy Speaker Shri Nanda Kishore Das (The New Ministry assumed charge of office on 1st April, 1937) 1. Hon'ble Captain Maharajah Sri Sri Sri Krishna . Prime Minister, Minister of Finance, Home Affairs, Chandra Gajapati Narayan Deo. Law and Commerce. 2. Hon'ble Shri Mandhata Gorachand Patnaik . Minister of Revenue and Education Mahasaya. 3. Hon'ble Maulavi Muhammad Latifur Rahman . Minister, Local Self Government and Public Works. (The Ministry resigned on the 19th July, 1937) (The New Ministry assumed charge of office on 19th July, 1937) As on 20th July, 1937 1. Hon'ble Shri Biswanath Das . Prime Minister, Minister of Finance, Home Affairs and Education. 2. Hon'ble Shri Nityananda Kanungo . Minister of Revenue and Public Works, [ Home w.e.f. 10th September 1937 ] 3. Hon'ble Shri Bodhram Dube . Minister of Law and Commerce, Health & Local Self- Government, [ Education w.e.f. 10th September, 1937] (The Ministry resigned on the 6th November, 1939) (The New Ministry assumed charge of office on 24th November, 1941) 1. Hon'ble Captain Maharajah Sri Sri Sri Krishna . Prime Minister, Minister of Home Affairs (excluding Chandra Gajapati Narayan Deo. Publicity), Local Self Government and Public Works 2. Hon'ble Pandit Godavaris Mishra . Minister of Finance, Home Affairs (Publicity), Development and Education 3. -

Biju Janata Dal Madan Mohan Bhuyan, Research Scolar, P.G
International Journal of Academic Research ISSN: 2348-7666 Vol.2, Issue-2(1), April-June, 2015 Sub- Regional Politics and Regional Political Parties: Biju Janata Dal Madan Mohan Bhuyan, Research Scolar, P.G. Department of History, Berhampur University, Berhampur-760007, Ganjam, Odisha Abstract: The new party Biju Janata Dal (BJD) had attracted a surprisingly large number of supporters, including 29 MLAs. Inherent contradictions and tensions among the Janata Dal Members of Legislative Assembly (MLAs), which had vitiated the political life in the state for last few months, after the demise of Biju Patnaik, came to an end with the formation of Biju Janata Dal in 1997. After the Lok Sabha election of 1998, in Odisha BJD-BJP combine became a major political force. This had resulted in the marginalization of the ruling Congress (I) and the decimation of the Janata Dal and its Left allies. Keywords: Biju Janata Dal, historical, geographical, vote bank Introduction Bharatiya Janata Party line for the While regionalism, in general, is party. Three senior leaders who were essentially a product of India‘s diversity, cabinet ministers in the previous Janata sub-regionalism in particular is the Dal government of Biju Patnaik- result of historical and geographical Biswabhusan Harichandan, Vedprakash factors in it. The term ‗sub-region‘ refers Agrawal, and Sarat Kar joined the to a smaller area within a region or Bharatiya Janata Party. A section of the ‗nation‘ which for economic geographic, state Janata Dal legislators had taken historical and social reasons is aware of enthusiastic interest to shape a joint possessing a distinct identity.(1) Sub- opposition front together with the regional conflicts have often been found Bharatiya Janata Party against the to take the shape of movements claiming congress in the fourth coming Odisha their separate identity in the formation Assembly election of 2000. -

Drainage System
CHAPTER DRAINAGE SYSTEM ou have observed water flowing through 2006) in this class . Can you, then, explain the the rivers, nalas and even channels reason for water flowing from one direction to Yduring rainy season which drain the the other? Why do the rivers originating from the excess water. Had these channels not been Himalayas in the northern India and the Western there, large-scale flooding would have Ghat in the southern India flow towards the east occurred. Wherever channels are ill-defined or and discharge their waters in the Bay of Bengal? choked, flooding is a common phenomenon. The flow of water through well-defined channels is known as ‘drainage’ and the network of such channels is called a ‘drainage system’. The drainage pattern of an area is the outcome of the geological time period, nature and structure of rocks, topography, slope, amount of water flowing and the periodicity of the flow. Do you have a river near your village or city? Have you ever been there for boating or bathing? Figure 3.1 : A River in the Mountainous Region Is it perennial (always with water) or ephemeral (water during rainy season, and dry, otherwise)? A river drains the water collected from a Do you know that rivers flow in the same specific area, which is called its ‘catchment area’. direction? You have studied about slopes in the An area drained by a river and its tributaries other two textbooks of geography (NCERT, is called a drainage basin. The boundary line Important Drainage Patterns (i) The drainage pattern resembling the branches of a tree is known as “dendritic” the examples of which are the rivers of northern plain. -

EXTRAORDINARY PUBLISHED by AUTHORITY No. 647, CUTTACK, SATURDAY, APRIL 21, 2018 / BAISAKHA 1, 1940 HOME (ELECTIONS) DEPAR
EXTRAORDINARY PUBLISHED BY AUTHORITY No. 647, CUTTACK, SATURDAY, APRIL 21, 2018 / BAISAKHA 1, 1940 HOME (ELECTIONS) DEPARTMENT NOTIFICATION The 3rd April, 2018 No. 3154– VE(A)-61/2018 /Elec.– The following Notification, dated the 12th March, 2018 of Election Commission of India, New Delhi is hereby republished in the Extraordinary Gazette of Odisha for general information. Sd/- SURENDRA KUMAR Chief Electoral Officer, Odisha ELECTION COMMISSION OF INDIA Nirvachan Sadan, Ashoka Road, New Delhi-110001 Dated 12 th March,2018 21, Phalguna, 1939 (Saka) NOTIFICATION No. 82/ECI/LET/TERR/ES-II/OR-LA/ (20 & 17/2014)/2018: - In pursuance of Section 106 of the Representation of the People Act, 1951 (43 of 1951), the Election Commission hereby published the Order of the High Court of Orissa, dated 30.01.2018 passed in Election Petition No. 20 of 2014 (Sri Sahadev Xaxa -Vrs- Jogesh Kumar Singh & others) and Election Petition No. 17 of 2014 (Sri Ajay Kumar Patel Vrs Sri Jogesh Kumar Singh & others related to the 9-Sundargarh (ST) Assembly Constituency. 2 HIGH COURT OF ORISSA: CUTTACK ELPET NO.20 OF 2014 & ELPET NO.17 OF 2014 In the matter of an application under Section 80 to 84 read with Sections 100,101 of the Representation of People Act, 1951 read with Orissa High Court Rules to regulate proceedings under the' Representation of People Act.1951. ………………….. ELPET NO.20 OF 2014 Sahadev Xaxa …. ….. Petitioner Versus Jogesh K. Singh & Others … … Respondents For Petitioner : M/s. Gopal Agarwal, K. K. Mishra & T. Mishra For opp. Parties : Mr. Pitambar Acharya, Senior Advocate. ELPET NO.17 OF 2014 Ajay Ku. -
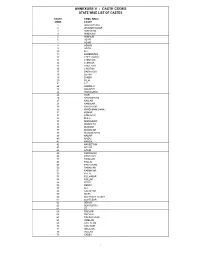
Annexure V - Caste Codes State Wise List of Castes
ANNEXURE V - CASTE CODES STATE WISE LIST OF CASTES STATE TAMIL NADU CODE CASTE 1 ADDI DIRVISA 2 AKAMOW DOOR 3 AMBACAM 4 AMBALAM 5 AMBALM 6 ASARI 7 ASARI 8 ASOOY 9 ASRAI 10 B.C. 11 BARBER/NAI 12 CHEETAMDR 13 CHELTIAN 14 CHETIAR 15 CHETTIAR 16 CRISTAN 17 DADA ACHI 18 DEYAR 19 DHOBY 20 DILAI 21 F.C. 22 GOMOLU 23 GOUNDEL 24 HARIAGENS 25 IYAR 26 KADAMBRAM 27 KALLAR 28 KAMALAR 29 KANDYADR 30 KIRISHMAM VAHAJ 31 KONAR 32 KONAVAR 33 M.B.C. 34 MANIGAICR 35 MOOPPAR 36 MUDDIM 37 MUNALIAR 38 MUSLIM/SAYD 39 NADAR 40 NAIDU 41 NANDA 42 NAVEETHM 43 NAYAR 44 OTHEI 45 PADAIACHI 46 PADAYCHI 47 PAINGAM 48 PALLAI 49 PANTARAM 50 PARAIYAR 51 PARMYIAR 52 PILLAI 53 PILLAIMOR 54 POLLAR 55 PR/SC 56 REDDY 57 S.C. 58 SACHIYAR 59 SC/PL 60 SCHEDULE CASTE 61 SCHTLEAR 62 SERVA 63 SOWRSTRA 64 ST 65 THEVAR 66 THEVAR 67 TSHIMA MIAR 68 UMBLAR 69 VALLALAM 70 VAN NAIR 71 VELALAR 72 VELLAR 73 YADEV 1 STATE WISE LIST OF CASTES STATE MADHYA PRADESH CODE CASTE 1 ADIWARI 2 AHIR 3 ANJARI 4 BABA 5 BADAI (KHATI, CARPENTER) 6 BAMAM 7 BANGALI 8 BANIA 9 BANJARA 10 BANJI 11 BASADE 12 BASOD 13 BHAINA 14 BHARUD 15 BHIL 16 BHUNJWA 17 BRAHMIN 18 CHAMAN 19 CHAWHAN 20 CHIPA 21 DARJI (TAILOR) 22 DHANVAR 23 DHIMER 24 DHOBI 25 DHOBI (WASHERMAN) 26 GADA 27 GADARIA 28 GAHATRA 29 GARA 30 GOAD 31 GUJAR 32 GUPTA 33 GUVATI 34 HARJAN 35 JAIN 36 JAISWAL 37 JASODI 38 JHHIMMER 39 JULAHA 40 KACHHI 41 KAHAR 42 KAHI 43 KALAR 44 KALI 45 KALRA 46 KANOJIA 47 KATNATAM 48 KEWAMKAT 49 KEWET 50 KOL 51 KSHTRIYA 52 KUMBHI 53 KUMHAR (POTTER) 54 KUMRAWAT 55 KUNVAL 56 KURMA 57 KURMI 58 KUSHWAHA 59 LODHI 60 LULAR 61 MAJHE -

ANSWERED ON:25.07.2016 Inclusion in ST List Gopal Dr
GOVERNMENT OF INDIA TRIBAL AFFAIRS LOK SABHA STARRED QUESTION NO:119 ANSWERED ON:25.07.2016 Inclusion in ST List Gopal Dr. K.;Majhi Shri Balabhadra Will the Minister of TRIBAL AFFAIRS be pleased to state: (a) The criteria and procedure being followed for inclusion/exclusion from and for carrying out other modifications in Scheduled Tribes (ST) List; (b) whether the Government has received proposals from various State Governments including Odisha for inclusion of various communities in the list of STs, and if so, the details thereof; (c) the action taken by the Government thereon along with the names of communities included in the list of STs during the last three years and the current year, State/UT-wise; (d) whether a number of proposals including those from Odisha for inclusion of tribes in the list of Scheduled Tribes are still pending with the Government for approval; and (e) if so, the details thereof and the reasons therefore along with the present status thereof and the time by which these proposals are likely to be approved, State/UT-wise? Answer MINISTER OF TRIBAL AFFAIRS (SHRI JUAL ORAM) (a) to (e) : A statement is laid on the table of the House. **** STATEMENT IN REFERENCE TO LOK SABHA STARRED QUESTION NO. 119 FOR 25-7-2016 REGARDING INCLUSION IN ST LIST BY SHRI BALABHADRA MAJHI AND DR. K. GOPAL. (a)Criteria followed for specification of a community as a Scheduled Tribe (ST) are: (i) indications of primitive traits, (ii) distinctive culture, (iii) geographical isolation, (iv) shyness of contact with the community at large, and (v) backwardness. -

Paper 18 History of Odisha
DDCE/History (M.A)/SLM/Paper-18 HISTORY OF ODISHA (FROM 1803 TO 1948 A.D.) By Dr. Manas Kumar Das CONTENT HISTORY OF ODISHA (From 1803 TO 1948 A.D.) Unit.No. Chapter Name Page No UNIT- I. a. British Occupation of Odisha. b. British Administration of Odisha: Land Revenue Settlements, administration of Justice. c. Economic Development- Agriculture and Industry, Trade and Commerce. UNIT.II. a. Resistance Movements in the 19th century- Khurda rising of 1804-05, Paik rebellion of 1817. b. Odisha during the revolt of 1857- role of Surendra Sai c. Tribal uprising- Ghumsar Rising under Dara Bisoi, Khond Rising under Chakra Bisoi, Bhuyan Rising Under Ratna Naik and Dharani Dhar Naik. UNIT – III. a. Growth of Modern Education, Growth of Press and Journalism. b. Natural Calamities in Odisha, Famine of 1866- its causes and effect. c. Social and Cultural changes in the 19th Century Odisha. d. Mahima Dharma. UNIT – IV. a. Oriya Movement: Growth of Socio-Political Associations, Growth of Public Associations in the 19th Century, Role of Utkal Sammilini (1903-1920) b. Nationalist Movement in Odisha: Non-Cooperation and Civil Disobedience Movements in Odisha. c. Creation of Separate province, Non-Congress and Congress Ministries( 1937-1947). d. Quit India Movement. e. British relation with Princely States of Odisha and Prajamandal Movement and Merger of the States. UNIT-1 Chapter-I British Occupation of Odisha Structure 1.1.0. Objectives 1.1.1. Introduction 1.1.2. British occupation of Odisha 1.1.2.1. Weakness of the Maratha rulers 1.1.2.2. Oppression of the land lords 1.1.2.3.