Depletion of the Third Complement Component Ameliorates Age- Dependent Oxidative Stress and Positively Modulates Autophagic Activity in Aged Retinas in a Mouse Model
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

To Study Mutant P53 Gain of Function, Various Tumor-Derived P53 Mutants
Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By Shama K Khokhar M.Sc., Bilaspur University, 2004 B.Sc., Bhopal University, 2002 2007 1 COPYRIGHT SHAMA K KHOKHAR 2007 2 WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES Date of Defense: 12-03-07 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY SHAMA KHAN KHOKHAR ENTITLED Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science Madhavi P. Kadakia, Ph.D. Thesis Director Daniel Organisciak , Ph.D. Department Chair Committee on Final Examination Madhavi P. Kadakia, Ph.D. Steven J. Berberich, Ph.D. Michael Leffak, Ph.D. Joseph F. Thomas, Jr., Ph.D. Dean, School of Graduate Studies 3 Abstract Khokhar, Shama K. M.S., Department of Biochemistry and Molecular Biology, Wright State University, 2007 Differential effect of TAp63γ mutants on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. p63, a member of the p53 gene family, known to play a role in development, has more recently also been implicated in cancer progression. Mice lacking p63 exhibit severe developmental defects such as limb truncations, abnormal skin, and absence of hair follicles, teeth, and mammary glands. Germline missense mutations of p63 have been shown to be responsible for several human developmental syndromes including SHFM, EEC and ADULT syndromes and are associated with anomalies in the development of organs of epithelial origin. -

Kallikrein 13: a New Player in Coronaviral Infections
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.01.971499; this version posted March 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Kallikrein 13: a new player in coronaviral infections. 2 3 Aleksandra Milewska1,2, Katherine Falkowski2, Magdalena Kalinska3, Ewa Bielecka3, 4 Antonina Naskalska1, Pawel Mak4, Adam Lesner5, Marek Ochman6, Maciej Urlik6, Jan 5 Potempa2,7, Tomasz Kantyka3,8, Krzysztof Pyrc1,* 6 7 1 Virogenetics Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian 8 University, Gronostajowa 7a, 30-387 Krakow, Poland. 9 2 Microbiology Department, Faculty of Biochemistry, Biophysics and Biotechnology, 10 Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland. 11 3 Laboratory of Proteolysis and Post-translational Modification of Proteins, Malopolska 12 Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, 13 Poland. 14 4 Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and 15 Biotechnology, Jagiellonian University, Gronostajowa 7 St., 30-387, Krakow, Poland. 16 5 University of Gdansk, Faculty of Chemistry, Wita Stwosza 63, 80-308 Gdansk, Poland. 17 6 Department of Cardiac, Vascular and Endovascular Surgery and Transplantology, Medical 18 University of Silesia in Katowice, Silesian Centre for Heart Diseases, Zabrze, Poland. 19 7 Centre for Oral Health and Systemic Diseases, University of Louisville School of Dentistry, 20 Louisville, KY 40202, USA. 21 8 Broegelmann Research Laboratory, Department of Clinical Science, University of Bergen, 22 5020 Bergen, Norway 23 24 25 26 27 28 29 30 31 * Correspondence should be addressed to Krzysztof Pyrc ([email protected]), Virogenetics 32 Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian University, 33 Gronostajowa 7, 30-387 Krakow, Poland; Phone number: +48 12 664 61 21; www: 34 http://virogenetics.info/. -
AACR2006-1686P
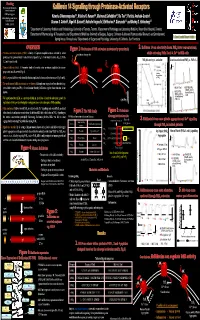
Funding Proteinases and Inflammation Network Group grant Kallikrein 14 Signalling through Proteinase-Activated Receptors CIHR operating grant 1,2 3,4 3,4 3,4 5 Alberta Heritage Foundation for Katerina Oikonomopoulou , Kristina K. Hansen , Mahmoud Saifeddine , Illa Tea , Patricia Andrade-Gordon , Medical Research Graeme S. Cottrell6, Nigel W. Bunnett6, Nathalie Vergnolle3, Eleftherios P. Diamandis1,2 and Morley D. Hollenberg3,4 NSERC / CRSNG 1Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto; 2Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto; 3Department of Pharmacology & Therapeutics, and 4Department of Medicine, University of Calgary, Calgary; 5Johnson & Johnson Pharmaceutical Research and Development, Spring House, Pennsylvania, 6Departments of Surgery and Physiology, University of California, San Francisco Kallikrein 14 can selectively disarm PAR (lower concentrations), OVERVIEW Figure 1: Mechanism of PAR activation (activation by proteolysis) 2. Kallikrein 14 can selectively disarm PAR1 (lower concentrations), 2+ Proteinase-activated receptors (PARs): a family of G-protein coupled receptors activated by serine proteinase cleavage site whilst activating PARs 2 and 4; Ca in HEK cells proteinases via a proteolytically revealed ‘tethered ligand’ (Fig. 1). Four family members (Fig. 2); PARs N PAR disarming 1, 2 and 4 signal to cells. N PAR1 dis-arming1 vs. activation Selective activation of PAR4 vs. PARs 1, 2 Human kallikreins (hKs): A 15-member family of secreted serine proteinases implicated in tumour 1 cm Thrombin 1 cm 2 min 2 min TFLLR-NH2 progression and cell survival (Fig. 4). (selective desensitization hK14: a tryptic kallikrein; wide tissue distribution, implicated in breast and ovarian cancer (Fig. 5 and 6). of PAR1) The mechanism of kallikrein action is not yet known: Although some targets have been identified (e.g. -

Aberrant Human Tissue Kallikrein Levels in the Stratum Corneum and Serum of Patients with Psoriasis: Dependence on Phenotype, Severity and Therapy N
CLINICAL AND LABORATORY INVESTIGATIONS DOI 10.1111/j.1365-2133.2006.07743.x Aberrant human tissue kallikrein levels in the stratum corneum and serum of patients with psoriasis: dependence on phenotype, severity and therapy N. Komatsu,* à K. Saijoh,§ C. Kuk,* F. Shirasaki,à K. Takeharaà and E.P. Diamandis* *Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario M5G 1L5, Canada àDepartment of Dermatology and §Department of Hygiene, Graduate School of Medical Science, School of Medicine, Kanazawa University, Kanazawa, Japan Summary Correspondence Background Human tissue kallikreins (KLKs) are a family of 15 trypsin-like or Eleftherios P. Diamandis. chymotrypsin-like secreted serine proteases (KLK1–KLK15). Multiple KLKs have E-mail: [email protected] been quantitatively identified in normal stratum corneum (SC) and sweat as can- didate desquamation-related proteases. Accepted for publication 7 November 2006 Objectives To quantify KLK5, KLK6, KLK7, KLK8, KLK10, KLK11, KLK13 and KLK14 in the SC and serum of patients with psoriasis, and their variation Key words between lesional and nonlesional areas and with phenotype, therapy and severity. diagnostic marker, human kallikreins, psoriasis, The overall SC serine protease activities were also measured. serine proteases, stratum corneum, therapy Methods Enzyme-linked immunosorbent assays and enzymatic assays were used. Conflicts of interest Results The lesional SC of psoriasis generally contained significantly higher levels None declared. of all KLKs. KLK6, KLK10 and KLK13 levels were significantly elevated even in the nonlesional SC. The overall trypsin-like, plasmin-like and furin-like activities were significantly elevated in the lesional SC. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Download, Or Email Articles for Individual Use
Florida State University Libraries Faculty Publications The Department of Biomedical Sciences 2010 Functional Intersection of the Kallikrein- Related Peptidases (KLKs) and Thrombostasis Axis Michael Blaber, Hyesook Yoon, Maria Juliano, Isobel Scarisbrick, and Sachiko Blaber Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] Article in press - uncorrected proof Biol. Chem., Vol. 391, pp. 311–320, April 2010 • Copyright ᮊ by Walter de Gruyter • Berlin • New York. DOI 10.1515/BC.2010.024 Review Functional intersection of the kallikrein-related peptidases (KLKs) and thrombostasis axis Michael Blaber1,*, Hyesook Yoon1, Maria A. locus (Gan et al., 2000; Harvey et al., 2000; Yousef et al., Juliano2, Isobel A. Scarisbrick3 and Sachiko I. 2000), as well as the adoption of a commonly accepted Blaber1 nomenclature (Lundwall et al., 2006), resolved these two fundamental issues. The vast body of work has associated 1 Department of Biomedical Sciences, Florida State several cancer pathologies with differential regulation or University, Tallahassee, FL 32306-4300, USA expression of individual members of the KLK family, and 2 Department of Biophysics, Escola Paulista de Medicina, has served to elevate the importance of the KLKs in serious Universidade Federal de Sao Paulo, Rua Tres de Maio 100, human disease and their diagnosis (Diamandis et al., 2000; 04044-20 Sao Paulo, Brazil Diamandis and Yousef, 2001; Yousef and Diamandis, 2001, 3 Program for Molecular Neuroscience and Departments of 2003; -

Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2008 Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES ACTIVATION PROFILES AND REGULATORY CASCADES OF THE HUMAN KALLIKREIN-RELATED PEPTIDASES By HYESOOK YOON A Dissertation submitted to the Department of Chemistry and Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Fall Semester, 2008 The members of the Committee approve the dissertation of Hyesook Yoon defended on July 10th, 2008. ________________________ Michael Blaber Professor Directing Dissertation ________________________ Hengli Tang Outside Committee Member ________________________ Brian Miller Committee Member ________________________ Oliver Steinbock Committee Member Approved: ____________________________________________________________ Joseph B. Schlenoff, Chair, Department of Chemistry and Biochemistry The Office of Graduate Studies has verified and approved the above named committee members. ii ACKNOWLEDGMENTS I would like to dedicate this dissertation to my parents for all your support, and my sister and brother. I would also like to give great thank my advisor, Dr. Blaber for his patience, guidance. Without him, I could never make this achievement. I would like to thank to all the members in Blaber lab. They are just like family to me and I deeply appreciate their kindness, consideration and supports. I specially like to thank to Mrs. Sachiko Blaber for her endless guidance and encouragement. I would like to thank Dr Jihun Lee, Margaret Seavy, Rani and Doris Terry for helpful discussions and supports. -

A Multiparametric Serum Kallikrein Panel for Diagnosis of Non ^ Small
Imaging, Diagnosis, Prognosis A Multiparametric Serum Kallikrein Panel for Diagnosis of Non ^ Small Cell Lung Carcinoma Chris Planque,1, 2 Lin Li,3 Yingye Zheng,3 Antoninus Soosaipillai,1, 2 Karen Reckamp,4 David Chia,5 Eleftherios P. Diamandis,1, 2 and Lee Goodglick5 Abstract Purpose: Human tissue kallikreins are a family of15 secreted serine proteases.We have previous- ly shown that the expression of several tissue kallikreins is significantly altered at the transcription- al level in lung cancer. Here, we examined the clinical value of 11members of the tissue kallikrein family as potential biomarkers for lung cancer diagnosis. Experimental Design: Serum specimens from 51 patients with non ^ small cell lung cancer (NSCLC) and from 50 healthy volunteers were collected. Samples were analyzed for11kallikreins (KLK1, KLK4-8, and KLK10-14) by specific ELISA. Data were statistically compared and receiver operating characteristic curves were constructed for each kallikrein and for various combinations. Results: Compared with sera from normal subjects, sera of patients with NSCLC had lower levels of KLK5, KLK7, KLK8, KLK10, and KLK12, and higher levels of KLK11, KLK13, and KLK14. Expres- sion of KLK11and KLK12 was positively correlated with stage.With the exception of KLK5, expres- sion of kallikreins was independent of smoking status and gender. KLK11, KLK12, KLK13, and KLK14 were associated with higher risk of NSCLC as determined by univariate analysis and con- firmed by multivariate analysis.The receiver operating characteristic curve of KLK4, KLK8, KLK10, KLK11,KLK12, KLK13, and KLK14 combined exhibited an area under the curve of 0.90 (95% con- fidence interval, 0.87-0.97). -

Natural and Synthetic Inhibitors of Kallikrein-Related Peptidases (Klks)
Biochimie 92 (2010) 1546e1567 Contents lists available at ScienceDirect Biochimie journal homepage: www.elsevier.com/locate/biochi Review Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs) Peter Goettig a,*, Viktor Magdolen b, Hans Brandstetter a a Division of Structural Biology, Department of Molecular Biology, University of Salzburg, Billrothstrasse 11, 5020 Salzburg, Austria b Klinische Forschergruppe der Frauenklinik, Klinikum rechts der Isar der TU München, Ismaninger Strasse 22, 81675 München, Germany article info abstract Article history: Including the true tissue kallikrein KLK1, kallikrein-related peptidases (KLKs) represent a family of fifteen Received 24 February 2010 mammalian serine proteases. While the physiological roles of several KLKs have been at least partially Accepted 29 June 2010 elucidated, their activation and regulation remain largely unclear. This obscurity may be related to the fact Available online 6 July 2010 that a given KLK fulfills many different tasks in diverse fetal and adult tissues, and consequently, the timescale of some of their physiological actions varies significantly. To date, a variety of endogenous þ Keywords: inhibitors that target distinct KLKs have been identified. Among them are the attenuating Zn2 ions, Tissue kallikrein fi active site-directed proteinaceous inhibitors, such as serpins and the Kazal-type inhibitors, or the huge, Speci city pockets fi Inhibitory compound unspeci c compartment forming a2-macroglobulin. Failure of these inhibitory systems can lead to certain Zinc pathophysiological conditions. One of the most prominent examples is the Netherton syndrome, which is Rule of five caused by dysfunctional domains of the Kazal-type inhibitor LEKTI-1 which fail to appropriately regulate KLKs in the skin. -

Supplementary Figure 1. Scatterplots Showing the Assignment of Total RNA-Seq Reads and Measures of Human Genes and Bacterial Species Detected in Each Sample
Supplementary Figure 1. Scatterplots showing the assignment of Total RNA-seq reads and measures of human genes and bacterial species detected in each sample. (A) Bacterial versus human-derived reads (millions). (B) Number of genes detected (minimum 20 assigned reads) versus human-derived reads (millions). (C) Number of bacterial species (minimum 20 assigned reads) versus bacterial-derived reads (millions). (D) Shannon diversity versus the number of bacterial-derived reads (millions). Supplementary Table 1. Patient #1 CT alignment CT gene CT protein SLIMM_all SLIMM_unique ompA Major Outer Membrane Protein 211 211 tmRNA noncoding RNA 173 173 omcA 9kDa-Cysteine-Rich Lipoprotein 100 100 omcB 60kDa Cysteine-Rich OMP 66 66 CT_398.1 noncoding RNA 61 61 groEL_1 HSP-60 39 39 ndk Nucleoside-2-P Kinase 33 33 CT_858 predicted Protease containing IRBP and DHR domains 29 29 tufA Elongation Factor Tu 28 28 hctB Histone-like Protein 2 24 24 yscC probable Yop proteins translocation protein C/general secretion pathway protein 23 23 CT_081 CHLTR T2 Protein 26 20 rpoC RNA Polymerase Beta' 24 20 CT_671 hypothetical protein 20 20 clpC ClpC Protease ATPase 19 19 CT_578 hypothetical protein 19 18 CT_664 adenylate cyclase-like protein 18 17 rho Transcription Termination Factor 15 15 glgA Glycogen Synthase 15 15 CT_456 hypothetical protein 14 14 CT_875 hypothetical protein 14 14 CT_708 SWF/SNF family helicase 13 13 dhnA Predicted 1%2C6-Fructose biphosphate aldolase (dehydrin family) 12 12 clpX CLP Protease ATPase 15 12 CT_082 hypothetical protein 13 11 nusA Transcription -

Human KLK13 / Kallikrein-13 Protein (His Tag)
Human KLK13 / Kallikrein-13 Protein (His Tag) Catalog Number: 10199-H08H General Information SDS-PAGE: Gene Name Synonym: KLK-L4; KLKL4 Protein Construction: A DNA sequence encoding the pro-form of human KLK13 (NP_056411.1) (Met 1-Ile 262) was expressed with a C-terminal polyhistidine tag. Source: Human Expression Host: HEK293 Cells QC Testing Purity: > 95 % as determined by SDS-PAGE Bio Activity: Protein Description Measured by its ability to cleave the fluorogenic peptide substrate Boc- VPR-AMC (R&D Systems, Catalog # ES011) . The specific activity is >200 Tissue kallikrein 13 (hK13), also known as KLK-L4 (kallikrein-like gene 4), pmoles/min/μg. (Activation description: The proenzyme needs to be is a member of the human tissue kallikrein family of serine proteases activated by Lysyl-Endopeptidase for an activated form) having diverse physiological functions in many tissues. The KLK13 gene resides on chromosome 19q13.3-4 along with other 14 members in a gene Endotoxin: cluster and shares a high degree of homology. KLK13 is a trypsin-like, secreted serine protease expressed specifically in the testicular tissue < 1.0 EU per μg of the protein as determined by the LAL method including prostate, salivary gland, breast, and testis. Growing evidence suggests that many kallikreins are implicated in carcinogenesis and may Stability: play a role in metastasis. KLK13 may be involved in the pathogenesis Samples are stable for up to twelve months from date of receipt at -70 ℃ and/or progression of breast and ovary cancers, and is regarded as a novel cancer biomarker. In addition, KLK13 interacts and forms complexes with Predicted N terminal: Gly 18 several serum protease inhibitors, such as alpha2-macroglobulin, and its expression is regulated by steroid hormones. -

Kallikreins Emerge As New Regulators of Viral Infections
Cellular and Molecular Life Sciences https://doi.org/10.1007/s00018-021-03922-7 Cellular andMolecular Life Sciences REVIEW Kallikreins emerge as new regulators of viral infections Georgios Pampalakis1 · Eleni Zingkou2 · Christos Panagiotidis1 · Georgia Sotiropoulou2 Received: 14 June 2021 / Revised: 23 July 2021 / Accepted: 12 August 2021 © The Author(s), under exclusive licence to Springer Nature Switzerland AG 2021 Abstract Kallikrein-related peptidases (KLKs) or kallikreins have been linked to diverse (patho) physiological processes, such as the epidermal desquamation and infammation, seminal clot liquefaction, neurodegeneration, and cancer. Recent mount- ing evidence suggests that KLKs also represent important regulators of viral infections. It is well-established that certain enveloped viruses, including infuenza and coronaviruses, require proteolytic processing of their hemagglutinin or spike proteins, respectively, to infect host cells. Similarly, the capsid protein of the non-enveloped papillomavirus L1 should be proteolytically cleaved for viral uncoating. Consequently, extracellular or membrane-bound proteases of the host cells are instrumental for viral infections and represent potential targets for drug development. Here, we summarize how extracellular proteolysis mediated by the kallikreins is implicated in the process of infuenza (and potentially coronavirus and papillo- mavirus) entry into host cells. Besides direct proteolytic activation of viruses, KLK5 and 12 promote viral entry indirectly through proteolytic cascade events, like the activation of thrombolytic enzymes that also can process hemagglutinin, while additional functions of KLKs in infection cannot be excluded. In the light of recent evidence, KLKs represent potential host targets for the development of new antivirals. Humanized animal models to validate their key functions in viral infections will be valuable.