CV Joseph Ryan, Ph.D
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Green Fluorescent Protein
P1: rpk/plb P2: rpk April 30, 1998 11:6 Annual Reviews AR057-17 Annu. Rev. Biochem. 1998. 67:509–44 Copyright c 1998 by Annual Reviews. All rights reserved THE GREEN FLUORESCENT PROTEIN Roger Y. Tsien Howard Hughes Medical Institute; University of California, San Diego; La Jolla, CA 92093-0647 KEY WORDS: Aequorea, mutants, chromophore, bioluminescence, GFP ABSTRACT In just three years, the green fluorescent protein (GFP) from the jellyfish Aequorea victoria has vaulted from obscurity to become one of the most widely studied and exploited proteins in biochemistry and cell biology. Its amazing ability to generate a highly visible, efficiently emitting internal fluorophore is both intrin- sically fascinating and tremendously valuable. High-resolution crystal structures of GFP offer unprecedented opportunities to understand and manipulate the rela- tion between protein structure and spectroscopic function. GFP has become well established as a marker of gene expression and protein targeting in intact cells and organisms. Mutagenesis and engineering of GFP into chimeric proteins are opening new vistas in physiological indicators, biosensors, and photochemical memories. CONTENTS NATURAL AND SCIENTIFIC HISTORY OF GFP .................................510 Discovery and Major Milestones .............................................510 Occurrence, Relation to Bioluminescence, and Comparison with Other Fluorescent Proteins .....................................511 PRIMARY, SECONDARY, TERTIARY, AND QUATERNARY STRUCTURE ...........512 Primary Sequence from -

Bioluminescence and Fluorescence of Three Sea Pens in the North-West
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.08.416396; this version posted December 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Bioluminescence and fluorescence of three sea pens in the north-west Mediterranean sea Warren R Francis* 1, Ana¨ısSire de Vilar 1 1: Dept of Biology, University of Southern Denmark, Odense, Denmark Corresponding author: [email protected] Abstract Bioluminescence of Mediterranean sea pens has been known for a long time, but basic parameters such as the emission spectra are unknown. Here we examined bioluminescence in three species of Pennatulacea, Pennatula rubra, Pteroeides griseum, and Veretillum cynomorium. Following dark adaptation, all three species could easily be stimulated to produce green light. All species were also fluorescent, with bioluminescence being produced at the same sites as the fluorescence. The shape of the fluorescence spectra indicates the presence of a GFP closely associated with light production, as seen in Renilla. Our videos show that light proceeds as waves along the colony from the point of stimulation for all three species, as observed in many other octocorals. Features of their bioluminescence are strongly suggestive of a \burglar alarm" function. Introduction Bioluminescence is the production of light by living organisms, and is extremely common in the marine environment [Haddock et al., 2010, Martini et al., 2019]. Within the phylum Cnidaria, biolumiescence is widely observed among the Medusazoa (true jellyfish and kin), but also among the Octocorallia, and especially the Pennatulacea (sea pens). -

The Diet and Predator-Prey Relationships of the Sea Star Pycnopodia Helianthoides (Brandt) from a Central California Kelp Forest
THE DIET AND PREDATOR-PREY RELATIONSHIPS OF THE SEA STAR PYCNOPODIA HELIANTHOIDES (BRANDT) FROM A CENTRAL CALIFORNIA KELP FOREST A Thesis Presented to The Faculty of Moss Landing Marine Laboratories San Jose State University In Partial Fulfillment of the Requirements for the Degree Master of Arts by Timothy John Herrlinger December 1983 TABLE OF CONTENTS Acknowledgments iv Abstract vi List of Tables viii List of Figures ix INTRODUCTION 1 MATERIALS AND METHODS Site Description 4 Diet 5 Prey Densities and Defensive Responses 8 Prey-Size Selection 9 Prey Handling Times 9 Prey Adhesion 9 Tethering of Calliostoma ligatum 10 Microhabitat Distribution of Prey 12 OBSERVATIONS AND RESULTS Diet 14 Prey Densities 16 Prey Defensive Responses 17 Prey-Size Selection 18 Prey Handling Times 18 Prey Adhesion 19 Tethering of Calliostoma ligatum 19 Microhabitat Distribution of Prey 20 DISCUSSION Diet 21 Prey Densities 24 Prey Defensive Responses 25 Prey-Size Selection 27 Prey Handling Times 27 Prey Adhesion 28 Tethering of Calliostoma ligatum and Prey Refugia 29 Microhabitat Distribution of Prey 32 Chemoreception vs. a Chemotactile Response 36 Foraging Strategy 38 LITERATURE CITED 41 TABLES 48 FIGURES 56 iii ACKNOWLEDGMENTS My span at Moss Landing Marine Laboratories has been a wonderful experience. So many people have contributed in one way or another to the outcome. My diving buddies perse- vered through a lot and I cherish our camaraderie: Todd Anderson, Joel Thompson, Allan Fukuyama, Val Breda, John Heine, Mike Denega, Bruce Welden, Becky Herrlinger, Al Solonsky, Ellen Faurot, Gilbert Van Dykhuizen, Ralph Larson, Guy Hoelzer, Mickey Singer, and Jerry Kashiwada. Kevin Lohman and Richard Reaves spent many hours repairing com puter programs for me. -

The Sandy Beach Environment
THE SANDY BEACH ENVIRONMENT The sandy beach is a region of shifting sands and crashing waves. Each time a wave pummels the shore, sediment is suspended and scattered in every direction. Together the wind and waves constantly rework and reshape the face of the beach. Throughout the year, the beach slope is continually changing. During the spring and summer, gentle constructive waves deposit sediment on the beach platform, building up the slope. The large destructive waves of fall and winter strip the sand from the beach, often leaving only cobblestones and the rocky beach platform. This unstable environment makes it difficult for plants and animals to settle. In addition to waves, the organisms must contend with the problems imposed by tidal fluctuations. The daily ebb and flow of the sea exposes the shore to all the elements. The animals living within the range of the high and low tides (intertidal) are subjected to extreme temperature changes, drying out, and other problems resulting from exposure to the air. Very few animals can survive these rigorous conditions, and no large plants can anchor in the shifting sands. To the casual observer, the sandy beach appears desolate and barren. However, within the sand grains live large numbers of diverse animals. To survive in this environment, these animals have evolved some interesting adaptations. Successful species either ride the waves, live just above the tideline, or burrow beneath the sand to protect themselves from the hammering waves. The upper beach -is full of small scavengers and beachhoppers. These animals live just above the breaking waves. -

An Invitation to Monitor Georgia's Coastal Wetlands
An Invitation to Monitor Georgia’s Coastal Wetlands www.shellfish.uga.edu By Mary Sweeney-Reeves, Dr. Alan Power, & Ellie Covington First Printing 2003, Second Printing 2006, Copyright University of Georgia “This book was prepared by Mary Sweeney-Reeves, Dr. Alan Power, and Ellie Covington under an award from the Office of Ocean and Coastal Resource Management, National Oceanic and Atmospheric Administration. The statements, findings, conclusions, and recommendations are those of the authors and do not necessarily reflect the views of OCRM and NOAA.” 2 Acknowledgements Funding for the development of the Coastal Georgia Adopt-A-Wetland Program was provided by a NOAA Coastal Incentive Grant, awarded under the Georgia Department of Natural Resources Coastal Zone Management Program (UGA Grant # 27 31 RE 337130). The Coastal Georgia Adopt-A-Wetland Program owes much of its success to the support, experience, and contributions of the following individuals: Dr. Randal Walker, Marie Scoggins, Dodie Thompson, Edith Schmidt, John Crawford, Dr. Mare Timmons, Marcy Mitchell, Pete Schlein, Sue Finkle, Jenny Makosky, Natasha Wampler, Molly Russell, Rebecca Green, and Jeanette Henderson (University of Georgia Marine Extension Service); Courtney Power (Chatham County Savannah Metropolitan Planning Commission); Dr. Joe Richardson (Savannah State University); Dr. Chandra Franklin (Savannah State University); Dr. Dionne Hoskins (NOAA); Dr. Charles Belin (Armstrong Atlantic University); Dr. Merryl Alber (University of Georgia); (Dr. Mac Rawson (Georgia Sea Grant College Program); Harold Harbert, Kim Morris-Zarneke, and Michele Droszcz (Georgia Adopt-A-Stream); Dorset Hurley and Aimee Gaddis (Sapelo Island National Estuarine Research Reserve); Dr. Charra Sweeney-Reeves (All About Pets); Captain Judy Helmey (Miss Judy Charters); Jan Mackinnon and Jill Huntington (Georgia Department of Natural Resources). -

CEAR Doc#961)
Vancouver Fraser Port Auihority PORT of i00 The Fcinie, 999 Canada Place ('Ð Varrcouyer. B.C. Canacia V6C 3T4 vancouver portvancouver.corn February 9,2018 Jocelyne Beaudet Panel Chair, Roberts Bank Terminal 2 Project C/O Debra Myles Panel Manager, Roberts Bank Terminal 2 Project Canadian Environmental Assessment Agency 22nd Floor, Place Bell 160 Elgin Street Ottawa, ON K1A 0H3 Dear Mme. Beaudet, From the Vancouver Fraser Port Authority re: Information Requests from the Review Panel for the Roberts Bank Terminal 2 Project Environmental Assessment: Responses (Select Responses from Packages 2 and 5 - February 9, 2018 Submission) The Vancouver Fraser Port Authority is pleased to submit to the Review Panel selected responses to Information Request Packages 2 and 5 related to the Roberts Bank Terminal 2 Project Environmental Impact Statement. We are making available the document Information Requests from the Review Panel for the Roberts Bank TermÌnal 2 Project Environmental Assessment: Responses (Se/ect Responses from Packages 2 and 5 - February 9,2078 Submission) which addresses the information requests IR2-19a, IR5-05, -06, -07, -1O, -45, and -47. Compilation of Panel Information Requests and Vancouver Fraser Port Authority Responses, which combines all available responses to the Review Panel's Information Requests, will be updated shortly While we will continue to regularly send responses as they are completed, we anticipate to complete Information Request Package 5 by March, Package 6 by April, and PackageT by June, and the Revised Project Description by May 2018. Submission of responses to Packages 8, 9 and 10 are still being planned and will be the subject of a further update to the Panel in the future. -

Hermit Crabs - Paguridae and Diogenidae
Identification Guide to Marine Invertebrates of Texas by Brenda Bowling Texas Parks and Wildlife Department April 12, 2019 Version 4 Page 1 Marine Crabs of Texas Mole crab Yellow box crab Giant hermit Surf hermit Lepidopa benedicti Calappa sulcata Petrochirus diogenes Isocheles wurdemanni Family Albuneidae Family Calappidae Family Diogenidae Family Diogenidae Blue-spot hermit Thinstripe hermit Blue land crab Flecked box crab Paguristes hummi Clibanarius vittatus Cardisoma guanhumi Hepatus pudibundus Family Diogenidae Family Diogenidae Family Gecarcinidae Family Hepatidae Calico box crab Puerto Rican sand crab False arrow crab Pink purse crab Hepatus epheliticus Emerita portoricensis Metoporhaphis calcarata Persephona crinita Family Hepatidae Family Hippidae Family Inachidae Family Leucosiidae Mottled purse crab Stone crab Red-jointed fiddler crab Atlantic ghost crab Persephona mediterranea Menippe adina Uca minax Ocypode quadrata Family Leucosiidae Family Menippidae Family Ocypodidae Family Ocypodidae Mudflat fiddler crab Spined fiddler crab Longwrist hermit Flatclaw hermit Uca rapax Uca spinicarpa Pagurus longicarpus Pagurus pollicaris Family Ocypodidae Family Ocypodidae Family Paguridae Family Paguridae Dimpled hermit Brown banded hermit Flatback mud crab Estuarine mud crab Pagurus impressus Pagurus annulipes Eurypanopeus depressus Rithropanopeus harrisii Family Paguridae Family Paguridae Family Panopeidae Family Panopeidae Page 2 Smooth mud crab Gulf grassflat crab Oystershell mud crab Saltmarsh mud crab Hexapanopeus angustifrons Dyspanopeus -

MARINE MAMMALS (MAMML1L.LEC UPDATE: February 5, 2013)
MARINE MAMMALS (MAMML1L.LEC UPDATE: February 5, 2013) - group of mammals highly specialized and adapted for completely aquatic existence - two groups not closely related, evolved from: * ungulates: Cetaceans * bears: Carnivora PHYLUM CHORDATA SUBPHYLUM VERTEBRATA CLASS MAMMALIA (hair, mammaries, homeotherms, viviparous) SUPERORDER CETACEA ENERGETIC CONSIDERATIONS - large size and homeothermy/endothermy * require large amounts of food for energy * energy transfer through food web at approx. 10% per step * must feed on low trophic level * must feed in predictably productive waters * requires migration for many species: benign breeding areas - upwelling: Sea of Cortez * abundant phytoplankton support zooplankton (crustaceans) and bait fishes - polar regions * ice plankton released in spring * supports large planktonic and benthic invert population * long days and unlimited nutrients * phytoplankton production swamps zooplankton and results in abundant detrital component ORDER MYSTICETI (baleen whales) - active suspension feeders use baleen filtering mechanism * comb-like plates replace teeth for filtering small organisms from water * plates for straining suspended from upper jaw in place of teeth * composed of keratin: tough flexible protein - baleen feeding mechanism * water drawn in and large tongue acts as piston to force out through baleen plates * food primarily small shoaling crustaceans called krill - three families: differences in coarseness and length of baleen - feeding types * water column * surface skim-feeding * bottom feeding -
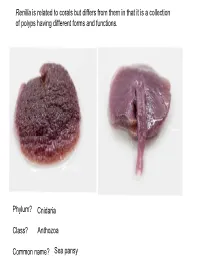
Phylum? Class? Common Name? Cnidaria Anthozoa Sea Pansy
Renilla is related to corals but differs from them in that it is a collection of polyps having different forms and functions. Phylum? Cnidaria Class? Anthozoa Common name? Sea pansy Phylum? Cnidaria Class? Hydrozoa Body form? Polyp Obelia colony Feeding polyp Phylum? Cnidaria Class? Hydrozoa Reproductive polyp Medusa bud – produced asexually Phylum? Cnidaria What is this? This is an Obelia medusa as seen under a compound microscope Class? Hydrozoa How does this reproduce? Sexually by producing eggs or sperm Phylum? Cnidaria Class? Hydrozoa What is the significance of the velum in the taxonomy of this organism? The velum is a characteristic of hydrozoan jellies. Thus Gonionemus is in Class Hydrozoa A B C D A: Exumbrella B: Subumbrella Goniomenus C: Manubrium D: Velum A: Tentacles C: Gonad A B B: Oral arm C Phylum? Cnidaria Identify Cells Class? Hydrozoa Physalia Tentacle Cnidocytes with nematocysts Phylum Cnidaria Class Hydrozoa 2. Identify structure within cell PhysaliaPhylum Tentacle Cnidaria Class Hydrozoa Nematocyst Physalia1. Identify Tentacle cell Cnidocytes with nematocysts Cnidocyte Phylum Cnidaria Class Hydrozoa Physalia Tentacle Cnidocytes with undischarged? Nematocysts Tentacle: Note Cnidocytes Bud (Asexual Reproduction) Phylum? Cnidaria Class Hydrozoa Hydra -Polyp Phylum Cnidaria Class Hydrozoa Obelia What stage of the lifecycle does this represent? Medusa stage (Sexually Reproduces) Phylum Cnidaria Class Hydrozoa Polpys Obelia Colony – A Colony of? Reproductive Polyp Asexual Stage Medusa Bud Feeding Polyp Phylum? Class? Common Name? Cnidaria Hydrozoa Portuguese Man-of-War Common Name? By-the-wind sailor or Velella Common name? Phylum? Class? A B Phylum? Cnidaria Class? Scyphozoa Common Name? Moon Jellly Phylum? Class? Common Name? Cnidaria Anthozoa Sea Anemone Kingdom? Animalia Phylum? Ctenophora Common name? Comb Jelly How does this animal move? Via cilia. -

Sea Pens and Burrowing Megafauna
SEA PENS AND BURROWING MEGAFAUNA An overview of dynamics and sensitivity characteristics for conservation management of marine SACs David J. Hughes Centre for Coastal and Marine Sciences Dunstaffnage Marine Laboratory, Oban AUGUST 1998 Prepared for Scottish Association for Marine Science (SAMS) UK Marine SACs Project, Task Manager A.M.W. Wilson, SAMS Acknowledgements I would like to thank the various reviewers of this report for their constructive suggestions and for access to unpublished information. Special thanks are due to Jim Atkinson, Mike Kaiser and Colin Chapman. I am also grateful to all others who provided information on particular sites, and to Jane Dodd and Elvira Poloczanska for their help with underwater photography. Citation: Hughes, D.J. 1998. Sea pens & burrowing megafauna (volume III). An overview of dynamics and sensitivity characteristics for conservation management of marine SACs. Scottish Association for Marine Science (UK Marine SACs Project). 105 Pages. CONTENTS PREFACE 5 EXECUTIVE SUMMARY 7 I. INTRODUCTION 13 A. NATURE AND IMPORTANCE OF THE BIOTOPE COMPLEX 13 B. KEY POINTS FROM CHAPTER I 23 II. STATUS AND DISTRIBUTION 25 A. STATUS WITHIN THE MNCR BIOTOPE CLASSIFICATION 25 B. OCCURRENCE WITHIN CANDIDATE SACs 26 C. DISTRIBUTION OUTSIDE THE BRITISH ISLES 35 D. KEY POINTS FROM CHAPTER II 36 III. ENVIRONMENTAL REQUIREMENTS AND PHYSICAL 37 ATTRIBUTES A. PHYSICAL ENVIRONMENT 37 B. KEY POINTS FROM CHAPTER III 41 IV. BIOLOGY AND ECOLOGICAL FUNCTIONING 43 A. BIOLOGY OF THE MAJOR CHARACTERIZING SPECIES 43 B. COMMUNITY ECOLOGY: INTERACTIONS BETWEEN SPECIES 46 C. KEY POINTS FROM CHAPTER IV 51 V. SENSITIVITY TO NATURAL EVENTS 53 A. CASE STUDIES OF POPULATION STABILITY AND CHANGE 53 B. -

Nudibranch Predators of Octocorallia Eric Brown Nova Southeastern University, [email protected]
Nova Southeastern University NSUWorks HCNSO Student Capstones HCNSO Student Work 4-29-2011 Nudibranch Predators of Octocorallia Eric Brown Nova Southeastern University, [email protected] This document is a product of extensive research conducted at the Nova Southeastern University . For more information on research and degree programs at the NSU , please click here. Follow this and additional works at: https://nsuworks.nova.edu/cnso_stucap Part of the Marine Biology Commons, and the Oceanography and Atmospheric Sciences and Meteorology Commons Share Feedback About This Item NSUWorks Citation Eric Brown. 2011. Nudibranch Predators of Octocorallia. Capstone. Nova Southeastern University. Retrieved from NSUWorks, . (23) https://nsuworks.nova.edu/cnso_stucap/23. This Capstone is brought to you by the HCNSO Student Work at NSUWorks. It has been accepted for inclusion in HCNSO Student Capstones by an authorized administrator of NSUWorks. For more information, please contact [email protected]. Nudibranch Predators of Octocorallia By Eric Brown A Capstone Review Paper Submitted in Partial Fulfillment of the Requirements for the Degree of Masters of Science: Marine Biology Eric Brown Nova Southeastern University Oceanographic Center April 2011 Capstone Committee Approval ______________________________ Dr. Joshua Feingold, Major Professor _____________________________ Dr. Charles Messing, Committee Member Table of Contents List of Figures ......................................................................................................................... -

Phylogeny and Systematics of Deep-Sea Sea Pens (Anthozoa: Octocorallia: Pennatulacea)
Molecular Phylogenetics and Evolution 69 (2013) 610-618 Contents lists available at ScienceDirect MOLECULAR PHYLOGENETICS & EVOLUTION Molecular Phylogenetics and Evolution ELSEVIER journal homepage: www.elsevier.com /locate/ym pev Phylogeny and systematics of deep-sea sea pens (Anthozoa: Octocorallia: Pennatulacea) Emily Dolana,b*, Paul A. Tyler3, Chris Yessonc, Alex D. Rogers' 1 aNational Oceanography Centre, University of Southampton, European Way, Southampton SOI4 3ZH, UK b Marine Biology Section, Biology Department, Ghent University, Krijgslaan 281 S8, Ghent B-9000, Belgium c Institute of Zoology, Zoological Society of London, Regent’s Park, London N W l 4RY, UK d Department of Zoology, University of Oxford, South Parks Road, Oxford 0X1 3PS, UK ARTICLE INFO ABSTRACT Article history: Molecular methods have been used for the first time to determine the phylogeny of families, genera and Received 5 November 2012 species within the Pennatulacea (sea pens). Variation in ND2 and mtMutS mitochondrial protein-coding Revised 4 July 2013 genes proved adequate to resolve phylogenetic relationships among pennatulacean families. The gene Accepted 19 July 2013 mtMutS is more variable than ND2 and differentiates all genera, and many pennatulacean species. A Available online 29 July 2013 molecular phylogeny based on a Bayesian analysis reveals that suborder Sessiliflorae is paraphyletic and Subselliflorae is polyphyletic. Many families of pennatulaceans do not represent monophyletic Keywords: groups including Umbellulidae, Pteroeididae, and Kophobelemnidae. The high frequency of morphologi Mitochondrial protein-coding genes Molecular systematics cal homoplasy in pennatulaceans has led to many misinterpretations in the systematics of the group. The mtMutS traditional classification scheme for pennatulaceans requires revision. ND2 © 2013 Elsevier Inc.