Convergent Evolution, Habitat Shifts and Variable Diversification Rates In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity and Structure of Bird and Mammal Communities in the Semiarid Chaco Region: Response to Agricultural Practices and Landscape Alterations
Diversity and structure of bird and mammal communities in the Semiarid Chaco Region: response to agricultural practices and landscape alterations Julieta Decarre March 2015 A thesis submitted for the degree of Doctor of Philosophy Division of Ecology and Evolution, Department of Life Sciences Imperial College London 2 Imperial College London Department of Life Sciences Diversity and structure of bird and mammal communities in the Semiarid Chaco Region: response to agricultural practices and landscape alterations Supervised by Dr. Chris Carbone Dr. Cristina Banks-Leite Dr. Marcus Rowcliffe Imperial College London Institute of Zoology Zoological Society of London 3 Declaration of Originality I herewith certify that the work presented in this thesis is my own and all else is referenced appropriately. I have used the first-person plural in recognition of my supervisors’ contribution. People who provided less formal advice are named in the acknowledgments. Julieta Decarre 4 Copyright Declaration The copyright of this thesis rests with the author and is made available under a Creative Commons Attribution Non-Commercial No Derivatives licence. Researchers are free to copy, distribute or transmit the thesis on the condition that they attribute it, that they do not use it for commercial purposes and that they do not alter, transform or build upon it. For any reuse or redistribution, researchers must make clear to others the licence terms of this work 5 “ …and we wandered for about four hours across the dense forest…Along the path I could see several footprints of wild animals, peccaries, giant anteaters, lions, and the footprint of a tiger, that is the first one I saw.” - Emilio Budin, 19061 I dedicate this thesis To my mother and my father to Virginia, Juan Martin and Alejandro, for being there through space and time 1 Book: “Viajes de Emilio Budin: La Expedición al Chaco, 1906-1907”. -

Habitat Fragmentation Compromises the Population Dynamic of the Globally Near-Threatened Straight-Billed Reedhaunter
bioRxiv preprint doi: https://doi.org/10.1101/468488; this version posted November 15, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Habitat fragmentation compromises the population dynamic of the globally near- 2 threatened Straight-billed Reedhaunter (Limnoctites rectirostris) 3 4 Maycon S. S. Gonçalves1,2, Priscila S. Pons1,2, Felipe C. Bonow2, Vinicius A. G. 5 Bastazini3, José A. Gil-Delgado1, Germán M. López-Iborra4 6 7 1Instituto Pró-Pampa (IPPampa), Laboratório de Ornitologia, Pelotas, Rio Grande do 8 Sul, Brazil 9 2Instituto Cavanilles de Biodiversidad y Biología Evolutiva, Universidad de Valencia, 10 Paterna, Valencia, Spain 11 3Theoretical and Experimental Ecological Station, French National Center for Scientific 12 Research and Paul Sabatier University, Moulis, France 13 4Departamento de Ecología - IMEM Ramon Margalef, Universidad de Alicante, 14 Alicante, Spain 15 16 Corresponding author: [email protected] 17 18 Abstract 19 Understanding the consequences of habitat fragmentation to biological populations is 20 crucial to develop sound conservation polices. The Straight-billed Reedhaunter 21 (Limnoctites rectirostris) is a little known and threatened Passeriform that is highly 22 dependent Erygo wetlands patches. Here, we evaluated the effects of habitat bioRxiv preprint doi: https://doi.org/10.1101/468488; this version posted November 15, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Lista Roja De Las Aves Del Uruguay 1
Lista Roja de las Aves del Uruguay 1 Lista Roja de las Aves del Uruguay Una evaluación del estado de conservación de la avifauna nacional con base en los criterios de la Unión Internacional para la Conservación de la Naturaleza. Adrián B. Azpiroz, Laboratorio de Genética de la Conservación, Instituto de Investigaciones Biológicas Clemente Estable, Av. Italia 3318 (CP 11600), Montevideo ([email protected]). Matilde Alfaro, Asociación Averaves & Facultad de Ciencias, Universidad de la República, Iguá 4225 (CP 11400), Montevideo ([email protected]). Sebastián Jiménez, Proyecto Albatros y Petreles-Uruguay, Centro de Investigación y Conservación Marina (CICMAR), Avenida Giannattasio Km 30.5. (CP 15008) Canelones, Uruguay; Laboratorio de Recursos Pelágicos, Dirección Nacional de Recursos Acuáticos, Constituyente 1497 (CP 11200), Montevideo ([email protected]). Cita sugerida: Azpiroz, A.B., M. Alfaro y S. Jiménez. 2012. Lista Roja de las Aves del Uruguay. Una evaluación del estado de conservación de la avifauna nacional con base en los criterios de la Unión Internacional para la Conservación de la Naturaleza. Dirección Nacional de Medio Ambiente, Montevideo. Descargo de responsabilidad El contenido de esta publicación es responsabilidad de los autores y no refleja necesariamente las opiniones o políticas de la DINAMA ni de las organizaciones auspiciantes y no comprometen a estas instituciones. Las denominaciones empleadas y la forma en que aparecen los datos no implica de parte de DINAMA, ni de las organizaciones auspiciantes o de los autores, juicio alguno sobre la condición jurídica de países, territorios, ciudades, personas, organizaciones, zonas o de sus autoridades, ni sobre la delimitación de sus fronteras o límites. -

Systematic Relationships and Biogeography of the Tracheophone Suboscines (Aves: Passeriformes)
MOLECULAR PHYLOGENETICS AND EVOLUTION Molecular Phylogenetics and Evolution 23 (2002) 499–512 www.academicpress.com Systematic relationships and biogeography of the tracheophone suboscines (Aves: Passeriformes) Martin Irestedt,a,b,* Jon Fjeldsaa,c Ulf S. Johansson,a,b and Per G.P. Ericsona a Department of Vertebrate Zoology and Molecular Systematics Laboratory, Swedish Museum of Natural History, P.O. Box 50007, SE-104 05 Stockholm, Sweden b Department of Zoology, University of Stockholm, SE-106 91 Stockholm, Sweden c Zoological Museum, University of Copenhagen, Copenhagen, Denmark Received 29 August 2001; received in revised form 17 January 2002 Abstract Based on their highly specialized ‘‘tracheophone’’ syrinx, the avian families Furnariidae (ovenbirds), Dendrocolaptidae (woodcreepers), Formicariidae (ground antbirds), Thamnophilidae (typical antbirds), Rhinocryptidae (tapaculos), and Conop- ophagidae (gnateaters) have long been recognized to constitute a monophyletic group of suboscine passerines. However, the monophyly of these families have been contested and their interrelationships are poorly understood, and this constrains the pos- sibilities for interpreting adaptive tendencies in this very diverse group. In this study we present a higher-level phylogeny and classification for the tracheophone birds based on phylogenetic analyses of sequence data obtained from 32 ingroup taxa. Both mitochondrial (cytochrome b) and nuclear genes (c-myc, RAG-1, and myoglobin) have been sequenced, and more than 3000 bp were subjected to parsimony and maximum-likelihood analyses. The phylogenetic signals in the mitochondrial and nuclear genes were compared and found to be very similar. The results from the analysis of the combined dataset (all genes, but with transitions at third codon positions in the cytochrome b excluded) partly corroborate previous phylogenetic hypotheses, but several novel arrangements were also suggested. -

TOUR REPORT Southwestern Amazonia 2017 Final
For the first time on a Birdquest tour, the Holy Grail from the Brazilian Amazon, Rondonia Bushbird – male (Eduardo Patrial) BRAZIL’S SOUTHWESTERN AMAZONIA 7 / 11 - 24 JUNE 2017 LEADER: EDUARDO PATRIAL What an impressive and rewarding tour it was this inaugural Brazil’s Southwestern Amazonia. Sixteen days of fine Amazonian birding, exploring some of the most fascinating forests and campina habitats in three different Brazilian states: Rondonia, Amazonas and Acre. We recorded over five hundred species (536) with the exquisite taste of specialties from the Rondonia and Inambari endemism centres, respectively east bank and west bank of Rio Madeira. At least eight Birdquest lifer birds were acquired on this tour: the rare Rondonia Bushbird; Brazilian endemics White-breasted Antbird, Manicore Warbling Antbird, Aripuana Antwren and Chico’s Tyrannulet; also Buff-cheeked Tody-Flycatcher, Acre Tody-Tyrant and the amazing Rufous Twistwing. Our itinerary definitely put together one of the finest selections of Amazonian avifauna, though for a next trip there are probably few adjustments to be done. The pre-tour extension campsite brings you to very basic camping conditions, with company of some mosquitoes and relentless heat, but certainly a remarkable site for birding, the Igarapé São João really provided an amazing experience. All other sites 1 BirdQuest Tour Report: Brazil’s Southwestern Amazonia 2017 www.birdquest-tours.com visited on main tour provided considerably easy and very good birding. From the rich east part of Rondonia, the fascinating savannas and endless forests around Humaitá in Amazonas, and finally the impressive bamboo forest at Rio Branco in Acre, this tour focused the endemics from both sides of the medium Rio Madeira. -
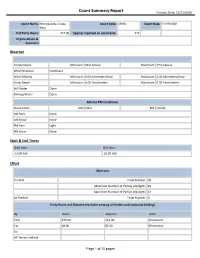
Count Summary Report Printout Date: 11/11/2016
Count Summary Report Printout Date: 11/11/2016 Count Name: Monteverde, Costa Count Code: CRMO Count Date: 12/19/2001 Rica # of Party Hours: 257.00 Species reported on count date: 376 Organizations & Sponsors: Weather Temperature Minimum: 59.0 Celsius Maximum: 77.0 Celsius Wind Direction Northeast Wind Velocity Minimum: 0.00 Kilometers/hour Maximum: 6.00 Kilometers/hour Snow Depth Minimum: 0.00 Centimeters Maximum: 0.00 Centimeters Still Water Open Moving Water Open AM and PM Conditions Cloud Cover AM: Clear PM: Cloudy AM Rain None AM Snow None PM Rain Light PM Snow None Start & End Times Start time End time 12:00 AM 10:00 AM Effort Observers In Field Total Number: 70 Minimum Number of Parties (daylight): 19 Maximum Number of Parties (daylight): 24 At Feeders Total Number: 0 Party Hours and Distance (excludes viewing at feeders and nocturnal birding) By Hours Distance Units Foot 239.00 114.00 Kilometers Car 18.00 85.00 Kilometers Air All-Terrain Vehicle Page 1 of 12 pages Count Summary Report Printout Date: 11/11/2016 Bicycle Dog Sled Golfcart Horseback Motorized Boat Non-Motorized Boat Skis/Xc-Skis Snowmachine Snowshoe Wheelchair Other Time and Distance Hours Distance Units At Feeders 0.00 Nocturnal Birding 17.00 26.00 Miles Total Party 257.00 199.00 Kilometers Checklist Species Number Number/Party Hrs. Flags Editorial Codes Highland Tinamou 3 0.0117 Great Tinamou 2 0.0078 Gray-headed Chachalaca 33 0.1284 Crested Guan 20 0.0778 Black Guan 18 0.0700 Black-breasted Wood-Quail 56 0.2179 Least Grebe 5 0.0195 Anhinga 1 0.0039 Fasciated Tiger-Heron -

Categorización De Las Aves De La Argentina
Categorización de las Aves de la Argentina SEGÚN SU ESTADO DE CONSERVACIÓN Informe del Ministerio de Ambiente y Desarrollo Sustentable de la Nación y de Aves Argentinas Ilustración: Leonardo González Galli - Gallito de arena AUTORIDADES Presidente de la Nación Mauricio Macri Ministro de Ambiente y Desarrollo Sustentable Sergio Bergman Jefa de Gabinete de Asesores Patricia Holzman Secretario de Política Ambiental, Cambio Climático y Desarrollo Sustentable Diego Moreno Subsecretaría de Planificación y Ordenamiento Ambiental del Territorio Dolores María Duverges Director Nacional de Biodiversidad y Recursos Hídricos Javier Garcia Espil Director de la Dirección de Fauna Silvestre y Conservación de la Biodiversidad Santiago D'Alessio 2 Indice CONTENIDO PRÓLOGO............................................................................................................................................5 INTRODUCCIÓN...................................................................................................................................7 METODOLOGÍA...................................................................................................................................9 2.1 Procedimientos generales y cambio de metodología...............................................9 2.2 Alcance geográfico para la recategorización.............................................................9 2.3 Elaboración de la matriz de especies y selección de especies para evaluar.........10 2.4. Proceso de evaluación y categorización de especies y justificación -

Scale, Pattern and Process in Biological Invasions
SCALE, PATTERN, AND PROCESS IN BIOLOGICAL INVASIONS By CRAIG R. ALLEN A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 1997 Copyright 1997 by Craig R. Allen ACKNOWLEDGEMENTS The work presented in this dissertation would not have been possible without the cooperation and encouragement of many. Foremost is the understanding of my immediate family, that is my wife Patty and now three-year-old son, Reece. Reece, while generally confused about what I was doing, nonetheless supported my effort to "write a book" in order to become a "doctor." Conflicts arose only when he needed my computer for dinosaur games. My co-advisors, W. M. Kitchens and C. S. Holling, encouraged my investigations and provided me with intellectual support and opportunity. For the same reasons, I extend my appreciation to my committee members, S. Humphrey, M. Moulton and D. Wojcik. Numerous friends and colleagues provided me with intellectual support and acted as a sounding board for ideas. Foremost are E. A. Forys, G. Peterson M. P. Moulton and J. Sendzemir as well as the entire "gang" of the Arthur Marshal Ecology Laboratory. I wish to thank all for their support and friendship. II! TABLE OF CONTENTS page ACKNOWLEDGEMENTS iii ABSTRACT viii INTRODUCTION 1 CHAPTERS 1. TRADITIONAL HYPOTHESES: INVASIONS AND EXTINCTIONS IN THE EVERGLADES ECOREGION 5 Introduction 5 Body-mass difference hypothesis 6 Diet difference hypothesis 7 Species replacement hypothesis 7 Phylogenetic hypothesis 8 Methods 8 Results 11 Discussion 14 2. LUMPY PATTERNS OF BODY MASS PREDICT INVASIONS AND EXTINCTIONS IN TRANSFORMING LANDSCAPES 18 Introduction 18 Methods and analysis 21 Species lists 21 Analysis 22 Results 26 Discussion 31 3. -

Bolivia: the Andes and Chaco Lowlands
BOLIVIA: THE ANDES AND CHACO LOWLANDS TRIP REPORT OCTOBER/NOVEMBER 2017 By Eduardo Ormaeche Blue-throated Macaw www.birdingecotours.com [email protected] 2 | T R I P R E P O R T Bolivia, October/November 2017 Bolivia is probably one of the most exciting countries of South America, although one of the less-visited countries by birders due to the remoteness of some birding sites. But with a good birding itinerary and adequate ground logistics it is easy to enjoy the birding and admire the outstanding scenery of this wild country. During our 19-day itinerary we managed to record a list of 505 species, including most of the country and regional endemics expected for this tour. With a list of 22 species of parrots, this is one of the best countries in South America for Psittacidae with species like Blue-throated Macaw and Red-fronted Macaw, both Bolivian endemics. Other interesting species included the flightless Titicaca Grebe, Bolivian Blackbird, Bolivian Earthcreeper, Unicolored Thrush, Red-legged Seriema, Red-faced Guan, Dot-fronted Woodpecker, Olive-crowned Crescentchest, Black-hooded Sunbeam, Giant Hummingbird, White-eared Solitaire, Striated Antthrush, Toco Toucan, Greater Rhea, Brown Tinamou, and Cochabamba Mountain Finch, to name just a few. We started our birding holiday as soon as we arrived at the Viru Viru International Airport in Santa Cruz de la Sierra, birding the grassland habitats around the terminal. Despite the time of the day the airport grasslands provided us with an excellent introduction to Bolivian birds, including Red-winged Tinamou, White-bellied Nothura, Campo Flicker, Chopi Blackbird, Chotoy Spinetail, White Woodpecker, and even Greater Rhea, all during our first afternoon. -

Divergence in Nest Placement and Parental Care of Neotropical Foliage‐Gleaners and Treehunters (Furnariidae: Philydorini)
J. Field Ornithol. 88(4):336–348, 2017 DOI: 10.1111/jofo.12227 Divergence in nest placement and parental care of Neotropical foliage-gleaners and treehunters (Furnariidae: Philydorini) Kristina L. Cockle,1,2,3,4 and Alejandro Bodrati2 1Instituto de Bio y Geociencias del NOA (IBIGEO-CONICET-UNSa), 9 de julio 14, Rosario de Lerma, Salta 4405, Argentina 2Proyecto Selva de Pino Parana, Velez Sarsfield y San Jurjo SN, San Pedro, Misiones 3352, Argentina 3Department of Forest and Conservation Sciences, University of British Columbia, 2424 Main Mall, Vancouver, British Columbia V6T 1Z4, Canada Received 25 July 2017; accepted 23 September 2017 ABSTRACT. The Neotropical ovenbirds (Furnariidae) are an adaptive radiation of suboscines renowned for the diversity of their nests. Like most altricial insectivores, they generally exhibit biparental care. One tribe, Philydorini, includes 46 species thought to nest in either underground burrows or tree cavities, nest types traditionally treated as equivalent in phylogenetic studies. Their parental care systems are poorly known, but could help illuminate how uniparental care – typically associated with frugivory – can arise in insectivores. We examined the extent to which nest placement, parental care, and associated reproductive traits map onto two major clades of Philydorini identified by genetic hypotheses. We review published literature and present new information from the Atlantic Forest of Argentina, including the first nest descriptions for Ochre-breasted Foliage-gleaners (Anabacerthia lichtensteini) and Sharp-billed Treehunters (Heliobletus contaminatus). In the Automolus-Thripadectes-Clibanornis clade (including Philydor rufum), 134 of 138 reported nests were in underground burrows. In the Syndactyla-Anabacerthia-Anabazenops clade (including Heliobletus, Philydor atricapillus, and Philydor erythrocercum), 44 of 48 nests were in tree cavities. -

Cooperative Breeding and Demography of Yellow Cardinal
ISSN (impISSNresso/printed) (printed) 0103-5657 ISSN (on-line) 2178-7875 Revista Brasileira de Ornitologia Volume 25 Issue 1 www.museu-goeldi.br/rbo March 2017 PublicadaPublished pela / Published by the by the Sociedade BrasileirBraziliana de Orn iOrnithologicaltologia / Brazil iSocietyan Ornithological Society RioBelém Grande - P -A RS ISSN (impresso/printed) 0103-5657 ISSN (on-line) 2178-7875 Revista Brasileira de Ornitologia Revista Brasileira EDITOR IN CHIEF Leandro Bugoni, Universidade Federal do Rio Grande - FURG, Rio Grande, RS E-mail: [email protected] MANAGING OFFICE ArVitortigos Moretti publicados and Regina na de R Siqueiraevista BuenoBrasileira de Ornitologia são indexados por: Biological Abstract, Scopus (Biobase, Geobase e EMBiology) e Zoological Record. de Ornitologia ASSOCIATE EDITORS Evolutionary Biology: Fábio Raposo do Amaral, Universidade Federal de São Paulo, Diadema, SP Manuscripts published by RevistaGustavo Br asileiSebastiánra Cabanne,de Ornitologia Museo Argentino are c odev eCienciasred b yNaturales the foll “Bernadinoowing indexingRivadavia”, Buenosdatabases: Aires, Argentina Biological Abstracts, ScopusJason D. (Weckstein,Biobase, Field Geobase, Museum ofand Natural EM HistoryBiology),, Chicago, and USA Zoological Records. Behavior: Carla Suertegaray Fontana, Pontifícia Universidade Católica do Rio Grande do Sul, Porto Alegre, RS Cristiano Schetini de Azevedo, Universidade Federal de Ouro Preto, Ouro Preto, MG Eduardo S. Santos, Universidade de São Paulo, São Paulo, SP Bibliotecas de referência para o depósito da versão impressa: Biblioteca do Museu de Zoologia Conservation:da USP, SP; Biblioteca doAlexander Museu Lees, N Manchesteracional, MetropolitanRJ; Biblioteca University do, Manchester, Museu UKParaense Emílio Goeldi, Ecology: PA; National Museum of CaioNatural Graco HMachado,istory UniversidadeLibrary, S Estadualmithsonian de Feira Idenstitution, Santana, Feira USA; de Santana, Louisiana BA State Systematics, Taxonomy,Universit andy, M Distribution:useum of NaturalLuciano Science, N. -

The Role of Size Assortment in Structuring Neotropical Bird Communities
Brooks, D.M. 2003. The role of size assortment in structuring Neotropical bird communities. Tx. J. Sci. 55: 59-74. THE ROLE OF SIZE ASSORTMENT IN STRUCTURING NEOTROPICAL BIRD COMMUNITIES Daniel M. Brooks Houston Museum of Natural Science; Department of Vertebrate Zoology; One Hermann Circle Dr.; Houston, Texas 77030-1799, USA ABSTRACT - I tested confamilial size assortment at three different latitudes, representing a gradient of productivity and stability: the northern subtropics (Rio Grande Valley), the equatorial zone (Amazonian Peru) and the austral subtropics (Paraguayan Chaco). Size assortment is the likely diminished persistence of a species by presence of morphologically similar species; temporally synchronous and spatially sympatric species competing for similar resources should exhibit distinct characters in ecomorphological space, molded over time to reduce the chance of competition. Despite least intensive sampling effort at the Amazon site, it is the most speciose (238 species, 78 common) compared to the Chaco (147, 76) and Rio Grande (61, 24) sites. Size assortment was tested by comparing mean mandibular measurements of confamilials in a real pool against those in a null pool. The pattern of size assortment was pervasive in 68% of the 22 families tested, with most being animal consumers or omnivores, represented by a high percentage of insectivores. EL PAPEL DE LA VARIEDAD DE TAMAÑO EN LA ESTRUCTURACIÓN DE LAS COMUNIDADES DE AVES NEOTROPICALES - La variedad del tamaño confamiliar (miembros de la misma familia) fue probada en tres latitudes diferentes representando un gradiente de productividad y estabilidad: el subtrópico septentrional (Valle del Río Grande), la zona ecuatorial (Amazonas peruano) y el subtrópico austral (Chaco paraguayo).