Poly(A)-Binding Proteins: Multifunctional Scaffolds for the Post-Transcriptional Control of Gene Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Large-Scale Analysis of Genome and Transcriptome Alterations in Multiple Tumors Unveils Novel Cancer-Relevant Splicing Networks
Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Research Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks Endre Sebestyén,1,5 Babita Singh,1,5 Belén Miñana,1,2 Amadís Pagès,1 Francesca Mateo,3 Miguel Angel Pujana,3 Juan Valcárcel,1,2,4 and Eduardo Eyras1,4 1Universitat Pompeu Fabra, E08003 Barcelona, Spain; 2Centre for Genomic Regulation, E08003 Barcelona, Spain; 3Program Against Cancer Therapeutic Resistance (ProCURE), Catalan Institute of Oncology (ICO), Bellvitge Institute for Biomedical Research (IDIBELL), E08908 L’Hospitalet del Llobregat, Spain; 4Catalan Institution for Research and Advanced Studies, E08010 Barcelona, Spain Alternative splicing is regulated by multiple RNA-binding proteins and influences the expression of most eukaryotic genes. However, the role of this process in human disease, and particularly in cancer, is only starting to be unveiled. We system- atically analyzed mutation, copy number, and gene expression patterns of 1348 RNA-binding protein (RBP) genes in 11 solid tumor types, together with alternative splicing changes in these tumors and the enrichment of binding motifs in the alter- natively spliced sequences. Our comprehensive study reveals widespread alterations in the expression of RBP genes, as well as novel mutations and copy number variations in association with multiple alternative splicing changes in cancer drivers and oncogenic pathways. Remarkably, the altered splicing patterns in several tumor types recapitulate those of undifferen- tiated cells. These patterns are predicted to be mainly controlled by MBNL1 and involve multiple cancer drivers, including the mitotic gene NUMA1. We show that NUMA1 alternative splicing induces enhanced cell proliferation and centrosome am- plification in nontumorigenic mammary epithelial cells. -

Downloaded from the TCGA Data Portal ( Data.Nci.Nih.Gov/Tcga/) (Supplemental Table S1)
bioRxiv preprint doi: https://doi.org/10.1101/023010; this version posted February 11, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks Endre Sebestyén1,*, Babita Singh1,*, Belén Miñana1,2, Amadís Pagès1, Francesca Mateo3, Miguel Angel Pujana3, Juan Valcárcel1,2,4, Eduardo Eyras1,4,5 1Universitat Pompeu Fabra, Dr. Aiguader 88, E08003 Barcelona, Spain 2Centre for Genomic Regulation, Dr. Aiguader 88, E08003 Barcelona, Spain 3Program Against Cancer Therapeutic Resistance (ProCURE), Catalan Institute of Oncology (ICO), Bellvitge Institute for Biomedical Research (IDIBELL), E08908 L’Hospitalet del Llobregat, Spain. 4Catalan Institution for Research and Advanced Studies, Passeig Lluís Companys 23, E08010 Barcelona, Spain *Equal contribution 5Correspondence to: [email protected] Keywords: alternative splicing, RNA binding proteins, splicing networks, cancer 1 bioRxiv preprint doi: https://doi.org/10.1101/023010; this version posted February 11, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Alternative splicing is regulated by multiple RNA-binding proteins and influences the expression of most eukaryotic genes. However, the role of this process in human disease, and particularly in cancer, is only starting to be unveiled. -

Mrna Turnover Philip Mitchell* and David Tollervey†
320 mRNA turnover Philip Mitchell* and David Tollervey† Nuclear RNA-binding proteins can record pre-mRNA are cotransported to the cytoplasm with the mRNP. These processing events in the structure of messenger proteins may preserve a record of the nuclear history of the ribonucleoprotein particles (mRNPs). During initial rounds of pre-mRNA in the cytoplasmic mRNP structure. This infor- translation, the mature mRNP structure is established and is mation can strongly influence the cytoplasmic fate of the monitored by mRNA surveillance systems. Competition for the mRNA and is used by mRNA surveillance systems that act cap structure links translation and subsequent mRNA as a checkpoint of mRNP integrity, particularly in the identi- degradation, which may also involve multiple deadenylases. fication of premature translation termination codons (PTCs). Addresses Cotransport of nuclear mRNA-binding proteins with mRNA Wellcome Trust Centre for Cell Biology, ICMB, University of Edinburgh, from the nucleus to the cytoplasm (nucleocytoplasmic shut- Kings’ Buildings, Edinburgh EH9 3JR, UK tling) was first observed for the heterogeneous nuclear *e-mail: [email protected] ribonucleoprotein (hnRNP) proteins. Some hnRNP proteins †e-mail: [email protected] are stripped from the mRNA at export [1], but hnRNP A1, Current Opinion in Cell Biology 2001, 13:320–325 A2, E, I and K are all exported (see [2]). Although roles for 0955-0674/01/$ — see front matter these hnRNP proteins in transport and translation have been © 2001 Elsevier Science Ltd. All rights reserved. reported [3•,4•], their affects on mRNA stability have been little studied. More is known about hnRNP D/AUF1 and Abbreviations AREs AU-rich sequence elements another nuclear RNA-binding protein, HuR, which act CBC cap-binding complex antagonistically to modulate the stability of a range of DAN deadenylating nuclease mRNAs containing AU-rich sequence elements (AREs) DSEs downstream sequence elements (reviewed in [2]). -

1 Title 1 Loss of PABPC1 Is Compensated by Elevated PABPC4
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.07.430165; this version posted February 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 1 Title 2 Loss of PABPC1 is compensated by elevated PABPC4 and correlates with transcriptome 3 changes 4 5 Jingwei Xie1, 2, Xiaoyu Wei1, Yu Chen1 6 7 1 Department of Biochemistry and Groupe de recherche axé sur la structure des 8 protéines, McGill University, Montreal, Quebec H3G 0B1, Canada 9 10 2 To whom correspondence should be addressed: Dept. of Biochemistry, McGill 11 University, Montreal, QC H3G 0B1, Canada. E-mail: [email protected]. 12 13 14 15 Abstract 16 Cytoplasmic poly(A) binding protein (PABP) is an essential translation factor that binds to 17 the 3' tail of mRNAs to promote translation and regulate mRNA stability. PABPC1 is the 18 most abundant of several PABP isoforms that exist in mammals. Here, we used the 19 CRISPR/Cas genome editing system to shift the isoform composition in HEK293 cells. 20 Disruption of PABPC1 elevated PABPC4 levels. Transcriptome analysis revealed that the 21 shift in the dominant PABP isoform was correlated with changes in key transcriptional 22 regulators. This study provides insight into understanding the role of PABP isoforms in 23 development and differentiation. 24 Keywords 25 PABPC1, PABPC4, c-Myc 26 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.07.430165; this version posted February 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins
Biomolecules 2015, 5, 1441-1466; doi:10.3390/biom5031441 OPEN ACCESS biomolecules ISSN 2218-273X www.mdpi.com/journal/biomolecules/ Article Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins Rebecca Bish 1,†, Nerea Cuevas-Polo 1,†, Zhe Cheng 1, Dolores Hambardzumyan 2, Mathias Munschauer 3, Markus Landthaler 3 and Christine Vogel 1,* 1 Center for Genomics and Systems Biology, Department of Biology, New York University, 12 Waverly Place, New York, NY 10003, USA; E-Mails: [email protected] (R.B.); [email protected] (N.C.-P.); [email protected] (Z.C.) 2 The Cleveland Clinic, Department of Neurosciences, Lerner Research Institute, 9500 Euclid Avenue, Cleveland, OH 44195, USA; E-Mail: [email protected] 3 RNA Biology and Post-Transcriptional Regulation, Max-Delbrück-Center for Molecular Medicine, Berlin-Buch, Robert-Rössle-Str. 10, Berlin 13092, Germany; E-Mails: [email protected] (M.M.); [email protected] (M.L.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-212-998-3976; Fax: +1-212-995-4015. Academic Editor: André P. Gerber Received: 10 May 2015 / Accepted: 15 June 2015 / Published: 15 July 2015 Abstract: DDX6 (p54/RCK) is a human RNA helicase with central roles in mRNA decay and translation repression. To help our understanding of how DDX6 performs these multiple functions, we conducted the first unbiased, large-scale study to map the DDX6-centric protein-protein interactome using immunoprecipitation and mass spectrometry. Using DDX6 as bait, we identify a high-confidence and high-quality set of protein interaction partners which are enriched for functions in RNA metabolism and ribosomal proteins. -

This Thesis Has Been Submitted in Fulfilment of the Requirements for a Postgraduate Degree (E.G
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: • This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. • A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. • This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. • The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. • When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Expression and subcellular localisation of poly(A)-binding proteins Hannah Burgess PhD The University of Edinburgh 2010 Abstract Poly(A)-binding proteins (PABPs) are important regulators of mRNA translation and stability. In mammals four cytoplasmic PABPs with a similar domain structure have been described - PABP1, tPABP, PABP4 and ePABP. The vast majority of research on PABP mechanism, function and sub-cellular localisation is however limited to PABP1 and little published work has explored the expression of PABP proteins. Here, I examine the tissue distribution of PABP1 and PABP4 in mouse and show that both proteins differ markedly in their expression at both the tissue and cellular level, contradicting the widespread perception that PABP1 is ubiquitously expressed. PABP4 is shown to be widely expressed though with an expression pattern distinct from PABP1, and thus may have a biological function in many tissues. -

Extensive Cross-Regulation of Post- Transcriptional Regulatory Networks in Drosophila
Downloaded from genome.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Extensive cross-regulation of post- transcriptional regulatory networks in Drosophila Marcus H. Stoibera,b, Sara Olsonc, Gemma E. Mayc, Michael O. Duffc, Jan Manentd, Robert Obard, K. G. Guruharshad,e, Spyros Artavanis-Tsakonasd,e,*, James B. Brownb,f,*, Brenton R. Graveleyc,* and Susan E. Celnikerb,* a Department of Biostatistics, University of California Berkeley, Berkeley, CA b Department of Genome Dynamics, Lawrence Berkeley National Laboratory, Berkeley, CA c Department of Genetics and Genome Sciences, Institute for Systems Genomics, University of Connecticut Health Center, Farmington, CT d Department of Cell Biology, Harvard Medical School, Boston, MA e Biogen Idec Inc, 14 Cambridge Center, Cambridge, MA 02142 USA f Department of Statistics, University of California Berkeley, Berkeley, CA * Corresponding Authors Susan Celniker, PhD Deputy of Science, Life Sciences Division Adjunct Professor, Comparative Biochemistry, University of California, Berkeley Co-Director Berkeley Drosophila Genome Project Lawrence Berkeley National Laboratory 1 Cyclotron Rd MS 977-180 Berkeley, CA 94720, USA E-mail: celniker @fruitfly.org Telephone: (510) 486-6258 Fax Number: (510) 495-2723 Brenton R. Graveley Department of Genetics and Genome Sciences Institute for Systems Genomics University of Connecticut Health Center Farmington, CT Email: [email protected] Phone: (860) 679-2090 Fax: (860) 679-8345 Downloaded from genome.cshlp.org on October 3, 2021 -

Mrna Export Through an Additional Cap-Binding Complex Consisting of NCBP1 and NCBP3
ARTICLE Received 2 Mar 2015 | Accepted 28 Jul 2015 | Published 18 Sep 2015 DOI: 10.1038/ncomms9192 OPEN mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3 Anna Gebhardt1,*, Matthias Habjan1,*, Christian Benda2, Arno Meiler1, Darya A. Haas1, Marco Y. Hein3, Angelika Mann1, Matthias Mann3, Bianca Habermann4 & Andreas Pichlmair1 The flow of genetic information from DNA to protein requires polymerase-II-transcribed RNA characterized by the presence of a 50-cap. The cap-binding complex (CBC), consisting of the nuclear cap-binding protein (NCBP) 2 and its adaptor NCBP1, is believed to bind all capped RNA and to be necessary for its processing and intracellular localization. Here we show that NCBP1, but not NCBP2, is required for cell viability and poly(A) RNA export. We identify C17orf85 (here named NCBP3) as a cap-binding protein that together with NCBP1 forms an alternative CBC in higher eukaryotes. NCBP3 binds mRNA, associates with components of the mRNA processing machinery and contributes to poly(A) RNA export. Loss of NCBP3 can be compensated by NCBP2 under steady-state conditions. However, NCBP3 becomes pivotal under stress conditions, such as virus infection. We propose the existence of an alternative CBC involving NCBP1 and NCBP3 that plays a key role in mRNA biogenesis. 1 Innate Immunity Laboratory, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. 2 Department of Structural Cell Biology, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. 3 Department of Proteomics and Signal Transduction, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. 4 Bioinformatics Core Facility, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. -

Structural and Functional Studies of Mrna Stability Regulators
Research Collection Doctoral Thesis Structural and Functional Studies of mRNA Stability Regulators Author(s): Ripin, Nina Publication Date: 2018-11 Permanent Link: https://doi.org/10.3929/ethz-b-000303696 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library DISS. ETH NO. 25327 Structural and functional studies of mRNA stability regulators A thesis submitted to attain the degree of DOCTOR OF SCIENCES of ETH ZÜRICH (Dr. sc. ETH Zürich) presented by NINA RIPIN Diplom-Biochemikerin, Goethe University, Frankfurt, Germany Born on 06.08.1986 citizen of Germany accepted on the recommendation of Prof. Dr. Frédéric Allain Prof. Dr. Stefanie Jonas Prof. Dr. Michael Sattler Prof. Dr. Witold Filipowicz 2018 “Success consists of going from failure to failure without loss of enthusiasm.” Winston Churchill Summary Posttranscriptional gene regulation (PTGR) is the process by which every step of the life cycle of an mRNA following transcription – maturation, transport, translation, subcellular localization and decay - is tightly regulated. This is accomplished by a complex network of multiple RNA binding proteins (RNPs) binding to several specific mRNA elements. Such cis-acting elements are or can be found within the 5’ cap, the 5’ untranslated region (UTR), the open reading frame (ORF), the 3’UTR and the poly(A) tail at the 3’ end of the mRNA. Adenylate-uridylate-rich elements (AU-rich elements; AREs) are heavily investigated regulatory cis- acting elements within 3’untranslated regions (3’UTRs). These are found in short-lived mRNAs and function as a signal for rapid degradation. -
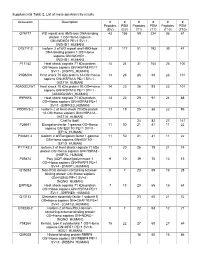
Supplemental Table 2: List of Mass-Spectrometry Results
Supplemental Table 2: List of mass-spectrometry results Accession Description # # # # # # Peptides PSM Peptides PSM Peptides PSM (EV) (EV) (T1) (T1) (T10) (T10) O75717 WD repeat and HMG-box DNA-binding 43 155 59 224 36 61 protein 1 OS=Homo sapiens GN=WDHD1 PE=1 SV=1 - [WDHD1_HUMAN] O75717-2 Isoform 2 of WD repeat and HMG-box 37 117 51 192 30 47 DNA-binding protein 1 OS=Homo sapiens GN=WDHD1 - [WDHD1_HUMAN] P11142 Heat shock cognate 71 kDa protein 14 24 31 104 26 100 OS=Homo sapiens GN=HSPA8 PE=1 SV=1 - [HSP7C_HUMAN] P0DMV8 Heat shock 70 kDa protein 1A OS=Homo 14 23 26 93 23 101 sapiens GN=HSPA1A PE=1 SV=1 - [HS71A_HUMAN] A0A0G2JIW1 Heat shock 70 kDa protein 1B OS=Homo 14 23 26 93 23 101 sapiens GN=HSPA1B PE=1 SV=1 - [A0A0G2JIW1_HUMAN] E9PKE3 Heat shock cognate 71 kDa protein 13 22 29 91 24 88 OS=Homo sapiens GN=HSPA8 PE=1 SV=1 - [E9PKE3_HUMAN] P0DMV8-2 Isoform 2 of Heat shock 70 kDa protein 13 19 25 86 22 95 1A OS=Homo sapiens GN=HSPA1A - [HS71A_HUMAN] Chaf1a (bait) 24 82 27 141 P26641 Elongation factor 1-gamma OS=Homo 11 50 21 81 11 22 sapiens GN=EEF1G PE=1 SV=3 - [EF1G_HUMAN] P26641-2 Isoform 2 of Elongation factor 1-gamma 11 50 21 81 11 22 OS=Homo sapiens GN=EEF1G - [EF1G_HUMAN] P11142-2 Isoform 2 of Heat shock cognate 71 kDa 11 20 26 79 21 74 protein OS=Homo sapiens GN=HSPA8 - [HSP7C_HUMAN] P09874 Poly [ADP-ribose] polymerase 1 9 10 39 70 13 15 OS=Homo sapiens GN=PARP1 PE=1 SV=4 - [PARP1_HUMAN] Q15233 Non-POU domain-containing octamer- 5 7 23 69 13 28 binding protein OS=Homo sapiens GN=NONO PE=1 SV=4 - [NONO_HUMAN] E9PNE6 Heat shock -

Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins?
International Journal of Molecular Sciences Review Messenger RNA Life-Cycle in Cancer Cells: Emerging Role of Conventional and Non-Conventional RNA-Binding Proteins? Lucie Coppin 1,2,3, Julie Leclerc 1,2,3, Audrey Vincent 1,2,3, Nicole Porchet 1,2,3 and Pascal Pigny 1,2,3,* 1 University of Lille, UMR-S 1172-JPARC—Jean-Pierre Aubert Research Center, F-59000 Lille, France; [email protected] (L.C.); [email protected] (J.L.); [email protected] (A.V.); [email protected] (N.P.) 2 Inserm, UMR-S 1172, Team “Mucins, Epithelial Differentiation and Carcinogenesis”, F-59000 Lille, Frances 3 CHU Lille, Service de Biochimie “Hormonologie, Métabolisme-Nutrition, Oncologie”, F-59000 Lille, France * Correspondence: [email protected]; Tel.: + 33-320-298-850 Received: 21 December 2017; Accepted: 19 February 2018; Published: 25 February 2018 Abstract: Functional specialization of cells and tissues in metazoans require specific gene expression patterns. Biological processes, thus, need precise temporal and spatial coordination of gene activity. Regulation of the fate of messenger RNA plays a crucial role in this context. In the present review, the current knowledge related to the role of RNA-binding proteins in the whole mRNA life-cycle is summarized. This field opens up a new angle for understanding the importance of the post-transcriptional control of gene expression in cancer cells. The emerging role of non-classic RNA-binding proteins is highlighted. The goal of this review is to encourage readers to view, through the mRNA life-cycle, novel aspects of the molecular basis of cancer and the potential to develop RNA-based therapies. -

PNAS 07-04849-SI Table 3. 6-18-2007
Table 5. β-arrestin 2-interacting proteins under nonstimulated (-) condition IPI accession Swiss- Number of Prot Gene symbol Protein name experiments accession number number detected Signal transduction Adaptor proteins IPI00027355 P32121 ARRB2 β-arrestin 2 6 IPI00293857 P49407 ARRB1 β-arrestin 1 6 IPI00021353 P10523 SAG S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin) 2 IPI00003917 P36575 ARR3 X-arrestin (Arrestin-C) (Cone arrestin) (cArr) (Retinal cone arrestin-3) (C-arrestin) 2 IPI00216318 P31946 YWHAB 14-3-3 β/α (14-3-3 protein beta/alpha) (Protein kinase C inhibitor protein 1) (KCIP-1) (Protein 1054) 2 IPI00220642 P61981 YWHAG 14-3-3 γ (14-3-3 protein gamma) (Protein kinase C inhibitor protein 1) (KCIP-1) 2 IPI00018146 P27348 YWHAQ 14-3-3 θ (14-3-3 protein tau) (14-3-3 protein theta) (14-3-3 protein T-cell) (HS1 protein) 2 IPI00216319 Q04917 YWHAH 14-3-3 η (14-3-3 protein eta) (Protein AS1) 2 IPI00000816 P62258 YWHAE 14-3-3 ε (14-3-3 protein epsilon) (14-3-3E) 2 Protein kinases IPI00027251 Q15208 STK38 STK38 (Serine/threonine-protein kinase 38) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1) 4 SCY1-like 2 (SCY1-like 2 protein) (coated vesicle-associated kinase of 104 kDa) (Eukaryotic protein IPI00396218 Q6P3W7 SCYL2 2 kinase family protein) (CDNA FLJ10074 fis, clone HEMBA1001744, weakly similar to SCY1 IPI00013835 Q13574 DGKZ DGK ζ (Diacylglycerol kinase zeta) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta) 2 DGK ε (Diacylglycerol kinase epsilon) (Diglyceride kinase epsilon) (DGK-epsilon)