Messenger-Rna-Binding Proteins and the Messages They Carry
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Ribosome As a Regulator of Mrna Decay
www.nature.com/cr www.cell-research.com RESEARCH HIGHLIGHT Make or break: the ribosome as a regulator of mRNA decay Anthony J. Veltri1, Karole N. D’Orazio1 and Rachel Green 1 Cell Research (2020) 30:195–196; https://doi.org/10.1038/s41422-019-0271-3 Cells regulate α- and β-tubulin levels through a negative present. To address this, the authors mixed pre-formed feedback loop which degrades tubulin mRNA upon detection TTC5–tubulin RNCs containing crosslinker with lysates from of excess free tubulin protein. In a recent study in Science, Lin colchicine-treated or colchicine-untreated TTC5-knockout cells et al. discover a role for a novel factor, TTC5, in recognizing (either having or lacking abundant free tubulin, respectively). the N-terminal motif of tubulins as they emerge from the After irradiation, TTC5 only crosslinked to the RNC in lysates ribosome and in signaling co-translational mRNA decay. from cells that had previously been treated with colchicine; Cells use translation-coupled mRNA decay for both quality these data suggested to the authors that some other (unknown) control and general regulation of mRNA levels. A variety of known factor may prevent TTC5 from binding under conditions of low quality control pathways including Nonsense Mediated Decay free tubulin. (NMD), No-Go Decay (NGD), and Non-Stop Decay (NSD) specifi- What are likely possibilities for how such coupling between cally detect and degrade mRNAs encoding potentially toxic translation and mRNA decay might occur? One example to protein fragments or sequences which cause ribosomes to consider is that of mRNA surveillance where extensive studies in translate poorly or stall.1 More generally, canonical mRNA yeast have identified a large group of proteins that recognize degradation is broadly thought to be translation dependent, and resolve stalled RNCs found on problematic mRNAs and 1234567890();,: though the mechanisms that drive these events are not target those mRNAs for decay. -

Coupling of Spliceosome Complexity to Intron Diversity
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.19.436190; this version posted March 20, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Coupling of spliceosome complexity to intron diversity Jade Sales-Lee1, Daniela S. Perry1, Bradley A. Bowser2, Jolene K. Diedrich3, Beiduo Rao1, Irene Beusch1, John R. Yates III3, Scott W. Roy4,6, and Hiten D. Madhani1,6,7 1Dept. of Biochemistry and Biophysics University of California – San Francisco San Francisco, CA 94158 2Dept. of Molecular and Cellular Biology University of California - Merced Merced, CA 95343 3Department of Molecular Medicine The Scripps Research Institute, La Jolla, CA 92037 4Dept. of Biology San Francisco State University San Francisco, CA 94132 5Chan-Zuckerberg Biohub San Francisco, CA 94158 6Corresponding authors: [email protected], [email protected] 7Lead Contact 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.19.436190; this version posted March 20, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. SUMMARY We determined that over 40 spliceosomal proteins are conserved between many fungal species and humans but were lost during the evolution of S. cerevisiae, an intron-poor yeast with unusually rigid splicing signals. We analyzed null mutations in a subset of these factors, most of which had not been investigated previously, in the intron-rich yeast Cryptococcus neoformans. -

Lin28b and Mir-142-3P Regulate Neuronal Differentiation by Modulating Staufen1 Expression
Cell Death and Differentiation (2018) 25, 432–443 & 2018 ADMC Associazione Differenziamento e Morte Cellulare All rights reserved 1350-9047/18 www.nature.com/cdd Lin28B and miR-142-3p regulate neuronal differentiation by modulating Staufen1 expression Younseo Oh1,5, Jungyun Park1,5, Jin-Il Kim1, Mi-Yoon Chang2, Sang-Hun Lee3, Youl-Hee Cho*,4 and Jungwook Hwang*,1,4 Staufen1 (STAU1) and Lin28B are RNA-binding proteins that are involved in neuronal differentiation as a function of post- transcriptional regulation. STAU1 triggers post-transcriptional regulation, including mRNA export, mRNA relocation, translation and mRNA decay. Lin28B also has multiple functions in miRNA biogenesis and the regulation of translation. Here, we examined the connection between STAU1 and Lin28B and found that Lin28B regulates the abundance of STAU1 mRNA via miRNA maturation. Decreases in the expression of both STAU1 and Lin28B were observed during neuronal differentiation. Depletion of STAU1 or Lin28B inhibited neuronal differentiation, and overexpression of STAU1 or Lin28B enhanced neuronal differentiation. Interestingly, the stability of STAU1 mRNA was modulated by miR-142-3p, whose maturation was regulated by Lin28B. Thus, miR-142-3p expression increased as Lin28B expression decreased during differentiation, leading to the reduction of STAU1 expression. The transcriptome from Staufen-mediated mRNA decay (SMD) targets during differentiation was analyzed, confirming that STAU1 was a key factor in neuronal differentiation. In support of this finding, regulation of STAU1 expression in mouse neural precursor cells had the same effects on neuronal differentiation as it did in human neuroblastoma cells. These results revealed the collaboration of two RNA-binding proteins, STAU1 and Lin28B, as a regulatory mechanism in neuronal differentiation. -

Ribosomes Slide on Lysine-Encoding Homopolymeric a Stretches
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Crossref RESEARCH ARTICLE elifesciences.org Ribosomes slide on lysine-encoding homopolymeric A stretches Kristin S Koutmou1, Anthony P Schuller1, Julie L Brunelle1,2, Aditya Radhakrishnan1, Sergej Djuranovic3, Rachel Green1,2* 1Department of Molecular Biology and Genetics, Johns Hopkins School of Medicine, Baltimore, United States; 2Howard Hughes Medical Institute, Johns Hopkins School of Medicine, Baltimore, United States; 3Department of Cell Biology and Physiology, Washington University School of Medicine, St. Louis, United States Abstract Protein output from synonymous codons is thought to be equivalent if appropriate tRNAs are sufficiently abundant. Here we show that mRNAs encoding iterated lysine codons, AAA or AAG, differentially impact protein synthesis: insertion of iterated AAA codons into an ORF diminishes protein expression more than insertion of synonymous AAG codons. Kinetic studies in E. coli reveal that differential protein production results from pausing on consecutive AAA-lysines followed by ribosome sliding on homopolymeric A sequence. Translation in a cell-free expression system demonstrates that diminished output from AAA-codon-containing reporters results from premature translation termination on out of frame stop codons following ribosome sliding. In eukaryotes, these premature termination events target the mRNAs for Nonsense-Mediated-Decay (NMD). The finding that ribosomes slide on homopolymeric A sequences explains bioinformatic analyses indicating that consecutive AAA codons are under-represented in gene-coding sequences. Ribosome ‘sliding’ represents an unexpected type of ribosome movement possible during translation. DOI: 10.7554/eLife.05534.001 *For correspondence: ragreen@ Introduction jhmi.edu Messenger RNA (mRNA) transcripts can contain errors that result in the production of incorrect protein products. -

Nuclear Expression of a Group II Intron Is Consistent with Spliceosomal Intron Ancestry
Downloaded from genesdev.cshlp.org on September 30, 2021 - Published by Cold Spring Harbor Laboratory Press Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry Venkata R. Chalamcharla, M. Joan Curcio, and Marlene Belfort1 Center for Medical Sciences, Wadsworth Center, New York State Department of Health, Albany, New York 12208, USA; and School of Public Health, State University of New York at Albany, Albany, New York 12201, USA Group II introns are self-splicing RNAs found in eubacteria, archaea, and eukaryotic organelles. They are mechanistically similar to the metazoan nuclear spliceosomal introns; therefore, group II introns have been invoked as the progenitors of the eukaryotic pre-mRNA introns. However, the ability of group II introns to function outside of the bacteria-derived organelles is debatable, since they are not found in the nuclear genomes of eukaryotes. Here, we show that the Lactococcus lactis Ll.LtrB group II intron splices accurately and efficiently from different pre-mRNAs in a eukaryote, Saccharomyces cerevisiae. However, a pre-mRNA harboring a group II intron is spliced predominantly in the cytoplasm and is subject to nonsense-mediated mRNA decay (NMD), and the mature mRNA from which the group II intron is spliced is poorly translated. In contrast, a pre-mRNA bearing the Tetrahymena group I intron or the yeast spliceosomal ACT1 intron at the same location is not subject to NMD, and the mature mRNA is translated efficiently. Thus, a group II intron can splice from a nuclear transcript, but RNA instability and translation defects would have favored intron loss or evolution into protein-dependent spliceosomal introns, consistent with the bacterial group II intron ancestry hypothesis. -
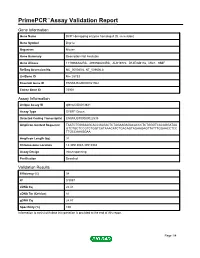
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DCP1 decapping enzyme homolog A (S. cerevisiae) Gene Symbol Dcp1a Organism Mouse Gene Summary Description Not Available Gene Aliases 1110066A22Rik, 4930568L04Rik, AU019772, D14Ertd817e, Mitc1, SMIF RefSeq Accession No. NC_000080.6, NT_039606.8 UniGene ID Mm.28733 Ensembl Gene ID ENSMUSG00000021962 Entrez Gene ID 75901 Assay Information Unique Assay ID qMmuCID0013841 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000022535 Amplicon Context Sequence TAATCTGGGAAGCACCGAGACTCTAGAAGAGACACCCTCTGGGTCACAGGATAA GTCTGCTCCGTCTGGTCATAAACATCTGACAGTAGAAGAGTTATTTGGAACCTCC TTGCCAAAGGAA Amplicon Length (bp) 91 Chromosome Location 14:30513043-30518984 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9997 cDNA Cq 22.41 cDNA Tm (Celsius) 81 gDNA Cq 24.87 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Dcp1a, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Exon Amplification: a Strategy to Isolate Mammalian Genes Based on RNA Splicing (Gene Cloning/Polymerase Chain Reaction) ALAN J
Proc. Natl. Acad. Sci. USA Vol. 88, pp. 4005-4009, May 1991 Genetics Exon amplification: A strategy to isolate mammalian genes based on RNA splicing (gene cloning/polymerase chain reaction) ALAN J. BUCKLER*, DAVID D. CHANG, SHARON L. GRAw, J. DAVID BROOK, DANIEL A. HABER, PHILLIP A. SHARP, AND DAVID E. HOUSMAN Center for Cancer Research, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139 Contributed by Phillip A. Sharp, January 25, 1991 ABSTRACT We have developed a method, exon amplifi- (7, 8). Thus, this method may be generally applicable for the cation, for fast and efficient isolation of coding sequences from selection of exon sequences from any gene. The method is complex mammalian genomic DNA. This method is based on also both rapid and easily adapted to large scale experiments. the selection of RNA sequences, exons, which are flanked by A series of cloned genomic DNA fragments can be screened functional 5' and 3' splice sites. Fragments of cloned genomic within 1-2 weeks. The sensitivity of this method is high. DNA are inserted into an intron, which is flanked by 5' and 3' Genomic DNA segments of 20 kilobases (kb) or more can be splice sites of the human immunodeficiency virus 1 tat gene successfully screened in a single transfection by using a set contained within the plasmid pSPL1. COS-7 cells are trans- of pooled subclones. This method thus allows the rapid fected with these constructs, and the resulting RNA transcripts identification of exons in mammalian genomic DNA and are processed in vivo. Splice sites of exons contained within the should facilitate the isolation of a wide spectrum of genes of inserted genomic fragment are paired with splice sites of the significance in physiology and development. -

Mrna Turnover Philip Mitchell* and David Tollervey†
320 mRNA turnover Philip Mitchell* and David Tollervey† Nuclear RNA-binding proteins can record pre-mRNA are cotransported to the cytoplasm with the mRNP. These processing events in the structure of messenger proteins may preserve a record of the nuclear history of the ribonucleoprotein particles (mRNPs). During initial rounds of pre-mRNA in the cytoplasmic mRNP structure. This infor- translation, the mature mRNP structure is established and is mation can strongly influence the cytoplasmic fate of the monitored by mRNA surveillance systems. Competition for the mRNA and is used by mRNA surveillance systems that act cap structure links translation and subsequent mRNA as a checkpoint of mRNP integrity, particularly in the identi- degradation, which may also involve multiple deadenylases. fication of premature translation termination codons (PTCs). Addresses Cotransport of nuclear mRNA-binding proteins with mRNA Wellcome Trust Centre for Cell Biology, ICMB, University of Edinburgh, from the nucleus to the cytoplasm (nucleocytoplasmic shut- Kings’ Buildings, Edinburgh EH9 3JR, UK tling) was first observed for the heterogeneous nuclear *e-mail: [email protected] ribonucleoprotein (hnRNP) proteins. Some hnRNP proteins †e-mail: [email protected] are stripped from the mRNA at export [1], but hnRNP A1, Current Opinion in Cell Biology 2001, 13:320–325 A2, E, I and K are all exported (see [2]). Although roles for 0955-0674/01/$ — see front matter these hnRNP proteins in transport and translation have been © 2001 Elsevier Science Ltd. All rights reserved. reported [3•,4•], their affects on mRNA stability have been little studied. More is known about hnRNP D/AUF1 and Abbreviations AREs AU-rich sequence elements another nuclear RNA-binding protein, HuR, which act CBC cap-binding complex antagonistically to modulate the stability of a range of DAN deadenylating nuclease mRNAs containing AU-rich sequence elements (AREs) DSEs downstream sequence elements (reviewed in [2]). -

Determinants of Exon 7 Splicing in the Spinal Muscular Atrophy Genes, SMN1 and SMN2 Luca Cartegni,*,† Michelle L
Determinants of Exon 7 Splicing in the Spinal Muscular Atrophy Genes, SMN1 and SMN2 Luca Cartegni,*,† Michelle L. Hastings,* John A. Calarco,‡ Elisa de Stanchina, and Adrian R. Krainer Cold Spring Harbor Laboratory, Cold Spring Harbor, NY Spinal muscular atrophy is a neurodegenerative disorder caused by the deletion or mutation of the survival-of- motor-neuron gene, SMN1. An SMN1 paralog, SMN2, differs by a CrT transition in exon 7 that causes substantial skipping of this exon, such that SMN2 expresses only low levels of functional protein. A better understanding of SMN splicing mechanisms should facilitate the development of drugs that increase survival motor neuron (SMN) protein levels by improving SMN2 exon 7 inclusion. In addition, exonic mutations that cause defective splicing give rise to many genetic diseases, and the SMN1/2 system is a useful paradigm for understanding exon-identity determinants and alternative-splicing mechanisms. Skipping of SMN2 exon 7 was previously attributed either to the loss of an SF2/ASF–dependent exonic splicing enhancer or to the creation of an hnRNP A/B–dependent exonic splicing silencer, as a result of the CrT transition. We report the extensive testing of the enhancer-loss and silencer- gain models by mutagenesis, RNA interference, overexpression, RNA splicing, and RNA-protein interaction ex- periments. Our results support the enhancer-loss model but also demonstrate that hnRNP A/B proteins antagonize SF2/ASF–dependent ESE activity and promote exon 7 skipping by a mechanism that is independent of the CrT transition and is, therefore, common to both SMN1 and SMN2. Our findings explain the basis of defective SMN2 splicing, illustrate the fine balance between positive and negative determinants of exon identity and alternative splicing, and underscore the importance of antagonistic splicing factors and exonic elements in a disease context. -

The Differential Interaction of Snrnps with Pre-Mrna Reveals Splicing Kinetics in Living Cells
Published October 4, 2010 This article has original data in the JCB Data Viewer JCB: Article http://jcb-dataviewer.rupress.org/jcb/browse/3011 The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells Martina Huranová,1 Ivan Ivani,1 Aleš Benda,2 Ina Poser,3 Yehuda Brody,4,5 Martin Hof,2 Yaron Shav-Tal,4,5 Karla M. Neugebauer,3 and David StanČk1 1Institute of Molecular Genetics and 2J. Heyrovský Institute of Physical Chemistry, Academy of Sciences of the Czech Republic, 142 20 Prague, Czech Republic 3Max Planck Institute for Molecular Cell Biology and Genetics, 01307 Dresden, Germany 4The Mina and Everard Goodman Faculty of Life Sciences and 5Institute for Nanotechnology and Advanced Materials, Bar-Ilan University, Ramat Gan 52900, Israel recursor messenger RNA (pre-mRNA) splicing is Core components of the spliceosome, U2 and U5 snRNPs, catalyzed by the spliceosome, a large ribonucleo- associated with pre-mRNA for 15–30 s, indicating that protein (RNP) complex composed of five small nuclear splicing is accomplished within this time period. Additionally, P Downloaded from RNP particles (snRNPs) and additional proteins. Using live binding of U1 and U4/U6 snRNPs with pre-mRNA oc- cell imaging of GFP-tagged snRNP components expressed curred within seconds, indicating that the interaction of at endogenous levels, we examined how the spliceosome individual snRNPs with pre-mRNA is distinct. These results assembles in vivo. A comprehensive analysis of snRNP are consistent with the predictions of the step-wise model dynamics in the cell nucleus enabled us to determine of spliceosome assembly and provide an estimate on the snRNP diffusion throughout the nucleoplasm as well as rate of splicing in human cells. -

A Upf3b-Mutant Mouse Model with Behavioral and Neurogenesis Defects
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Mol Psychiatry Manuscript Author . Author Manuscript Author manuscript; available in PMC 2018 September 27. Published in final edited form as: Mol Psychiatry. 2018 August ; 23(8): 1773–1786. doi:10.1038/mp.2017.173. A Upf3b-mutant mouse model with behavioral and neurogenesis defects L Huang1, EY Shum1, SH Jones1, C-H Lou1, J Dumdie1, H Kim1, AJ Roberts2, LA Jolly3,4, J Espinoza1, DM Skarbrevik1, MH Phan1, H Cook-Andersen1, NR Swerdlow5, J Gecz3,4, and MF Wilkinson1,6 1Department of Reproductive Medicine, School of Medicine, University of California, San Diego, La Jolla, California, USA 2Department of Molecular and Cellular Neuroscience, The Scripps Research Institute, 10550 North Torrey Pines Road, MB6, La Jolla, CA 92037, USA 3School Adelaide Medical School and Robison Research Institute, University of Adelaide, Adelaide, SA 5005, Australia 4South Australian Health and Medical Research Institute, Adelaide, SA, 5005, Australia 5Department of Psychiatry, School of Medicine, University of California, San Diego, La Jolla, California, USA 6Institute of Genomic Medicine, University of California, San Diego, La Jolla, CA Abstract Nonsense-mediated RNA decay (NMD) is a highly conserved and selective RNA degradation pathway that acts on RNAs terminating their reading frames in specific contexts. NMD is regulated in a tissue-specific and developmentally controlled manner, raising the possibility that it influences developmental events. Indeed, loss or depletion of NMD factors have been shown to disrupt developmental events in organisms spanning the phylogenetic scale. In humans, mutations in the NMD factor gene, UPF3B, cause intellectual disability (ID) and are strongly associated with autism spectrum (ASD), attention deficit hyperactivity disorder (ADHD), and schizophrenia (SCZ). -

Dissertation
Regulation of gene silencing: From microRNA biogenesis to post-translational modifications of TNRC6 complexes DISSERTATION zur Erlangung des DOKTORGRADES DER NATURWISSENSCHAFTEN (Dr. rer. nat.) der Fakultät Biologie und Vorklinische Medizin der Universität Regensburg vorgelegt von Johannes Danner aus Eggenfelden im Jahr 2017 Das Promotionsgesuch wurde eingereicht am: 12.09.2017 Die Arbeit wurde angeleitet von: Prof. Dr. Gunter Meister Johannes Danner Summary ‘From microRNA biogenesis to post-translational modifications of TNRC6 complexes’ summarizes the two main projects, beginning with the influence of specific RNA binding proteins on miRNA biogenesis processes. The fate of the mature miRNA is determined by the incorporation into Argonaute proteins followed by a complex formation with TNRC6 proteins as core molecules of gene silencing complexes. miRNAs are transcribed as stem-loop structured primary transcripts (pri-miRNA) by Pol II. The further nuclear processing is carried out by the microprocessor complex containing the RNase III enzyme Drosha, which cleaves the pri-miRNA to precursor-miRNA (pre-miRNA). After Exportin-5 mediated transport of the pre-miRNA to the cytoplasm, the RNase III enzyme Dicer cleaves off the terminal loop resulting in a 21-24 nt long double-stranded RNA. One of the strands is incorporated in the RNA-induced silencing complex (RISC), where it directly interacts with a member of the Argonaute protein family. The miRNA guides the mature RISC complex to partially complementary target sites on mRNAs leading to gene silencing. During this process TNRC6 proteins interact with Argonaute and recruit additional factors to mediate translational repression and target mRNA destabilization through deadenylation and decapping leading to mRNA decay.