Microbiome Assembly of Avian Eggshells and Their Potential As Transgenerational Carriers of Maternal Microbiota
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nest Survival in Year-Round Breeding Tropical Red-Capped Larks
University of Groningen Nest survival in year-round breeding tropical red-capped larks Calandrella cinerea increases with higher nest abundance but decreases with higher invertebrate availability and rainfall Mwangi, Joseph; Ndithia, Henry K.; Kentie, Rosemarie; Muchai, Muchane; Tieleman, B. Irene Published in: Journal of Avian Biology DOI: 10.1111/jav.01645 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2018 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Mwangi, J., Ndithia, H. K., Kentie, R., Muchai, M., & Tieleman, B. I. (2018). Nest survival in year-round breeding tropical red-capped larks Calandrella cinerea increases with higher nest abundance but decreases with higher invertebrate availability and rainfall. Journal of Avian Biology, 49(8), [01645]. https://doi.org/10.1111/jav.01645 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. -

Avian Adaptation Along an Aridity Gradient. Physiology, Behavior, And
AVIAN ADAPTATION ALONG AN ARIDITY GRADIENT PHYSIOLOGY, BEHAVIOR, AND LIFE HISTORY B. Irene Tieleman This research was financially supported by Schuurman Schimmel van Outeren Stichting National Wildlife Research Center, Taif, Saudi Arabia Schure Beijerinck Popping Fonds Lay-out: Heerko Tieleman Figures: Dick Visser Photographs: Irene Tieleman © 2002 Irene Tieleman ISBN-nummer: 90-367-1726-4 Electronic version 90-367-1727-2 RIJKSUNIVERSITEIT GRONINGEN Avian adaptation along an aridity gradient physiology, behavior, and life history Proefschrift ter verkrijging van het doctoraat in de Wiskunde en Natuurwetenschappen aan de Rijksuniversiteit Groningen op gezag van de Rector Magnificus, dr. F. Zwarts, in het openbaar te verdedigen op dinsdag 10 december 2002 om 13.15 uur door Bernadine Irene Tieleman geboren op 15 juni 1973 te Groningen Promotores: Prof. S. Daan Prof. J.B. Williams Beoordelingscommissie: Prof. W.R. Dawson Prof. R.H. Drent Prof. R.E. Ricklefs Contents PART I: INTRODUCTION 1. General introduction 11 2. Physiological ecology and behavior of desert birds 19 3. The adjustment of avian metabolic rates and water fluxes to desert 61 environments PART II: PHYSIOLOGY AND BEHAVIOR OF LARKS ALONG AN ARIDITY GRADIENT 4. Adaptation of metabolism and evaporative water loss along an 89 aridity gradient 5. Phenotypic variation of larks along an aridity gradient: 105 are desert birds more flexible? 6. Physiological adjustments to arid and mesic environments in larks 131 (Alaudidae) 7. Cutaneous and respiratory water loss in larks from arid and 147 mesic environments 8. Energy and water budgets of larks in a life history perspective: 165 is parental effort related to environmental aridity? PART III: PHYSIOLOGICAL MECHANISMS 9. -

Identification of Oriental Skylark
Identification of Oriental Skylark Hadoram Shirihai he Oriental Skylark Alauda gulgula (also sometimes known as Small TSkylark, Lesser Skylark or Eastern Skylark) is found across a large area of southern Asia. Eleven races were recognised by Vaurie (1959), most of which are resident in tropical Asia, but A. g. inconspicua, which breeds west to central Asia and Iran, is migratory, though its winter quarters are not known. The species has not yet been reliably recorded in Europe, although there are several recent records for Israel (see final section, and Shirihai in prep.) and it is possible that the Oriental Skylark will eventually be found in western Europe, and perhaps even Britain and Ireland. The main confusion species is Skylark A. arvensis, especially the smaller races. Given good views, however, the careful observer should not find separating them a serious problem. This paper summarises the main identification features of Oriental Skylark and its distinction from Skylark and other larks. Identification in the field In the field, Oriental Skylark resembles Skylark in coloration, but Wood- lark Lullula arborea in shape and flight. Its pointed bill is relatively long and thick, and it has a shortish tail and relatively long legs. From a distance, it might even be confused with Short-toed Calandrella brachydactyla or Lesser Short-toed Lark C. rufescens. The following are important points to observe when identifying the Oriental Skylark in the field. SILHOUETTE AND SIZE Size as Woodlark lark's. Wings rather short, primaries project- (about 16 cm in length), significantly smaller ing little, if at all, beyond tertials, unlike than nominate Skylark (18.5 cm). -

Western Birds, Index, 2000–2009
WESTERN BIRDS, INDEX, 2000–2009 Volumes 31 (2000), 32 (2001), 33 (2002), 34 (2003), 35 (2004), 36 (2005), 37 (2006), 38 (2007), 39 (2008), and 40 (2009) Compiled by Daniel D. Gibson abeillei, Icterus bullockii—38:99 acadicus, Aegolius acadicus—36:30; 40:98 Accentor, Siberian—31:57; 36:38, 40, 50–51 Accipiter cooperii—31:218; 33:34–50; 34:66, 207; 35:83; 36:259; 37:215–227; 38:133; 39:202 gentilis—35:112; 39:194; 40:78, 128 striatus—32:101, 107; 33:18, 34–50; 34:66; 35:108–113; 36:196; 37:12, 215–227; 38:133; 40:78, 128 Acevedo, Marcos—32:see Arnaud, G. aciculatus, Agelaius phoeniceus—35:229 Acridotheres javanicus—34:123 Acrocephalus schoenobaenus—39:196 actia, Eremophila alpestris—36:228 Actitis hypoleucos—36:49 macularia—32:108, 145–166; 33:69–98, 134–174, 222–240; 34:68 macularius—35:62–70, 77–87, 186, 188, 194–195; 36:207; 37:1–7, 12, 34; 40:81 acuflavidus, Thalasseus sandvicensis—40:231 adastus, Empidonax traillii—32:37; 33:184; 34:125; 35:197; 39:8 Aechmophorus clarkii—34:62, 133–148; 36:144–145; 38:104, 126, 132 occidentalis—34:62, 133–148; 36:144, 145, 180; 37:34; 38:126; 40:58, 75, 132–133 occidentalis/clarkii—34:62 (sp.)—35:126–146 Aegolius acadicus—32:110; 34:72, 149–156; 35:176; 36:30, 303–309; 38:107, 115–116; 40:98 funereus—36:30; 40:98 Aeronautes montivagus—34:207 saxatalis—31:220; 34:73, 186–198, 199–203, 204–208, 209–215, 216–224, 245; 36:218; 37:29, 35, 149–155; 38:82, 134, 261–267 aestiva, Dendroica petechia—40:297 Aethia cristatella—36:29; 37:139–148, 197, 199, 210 psittacula—31:14; 33:1, 14; 34:163; 36:28; 37:95, 139, -

Multilocus Phylogeny of the Avian Family Alaudidae (Larks) Reveals
1 Multilocus phylogeny of the avian family Alaudidae (larks) 2 reveals complex morphological evolution, non- 3 monophyletic genera and hidden species diversity 4 5 Per Alströma,b,c*, Keith N. Barnesc, Urban Olssond, F. Keith Barkere, Paulette Bloomerf, 6 Aleem Ahmed Khang, Masood Ahmed Qureshig, Alban Guillaumeth, Pierre-André Crocheti, 7 Peter G. Ryanc 8 9 a Key Laboratory of Zoological Systematics and Evolution, Institute of Zoology, Chinese 10 Academy of Sciences, Chaoyang District, Beijing, 100101, P. R. China 11 b Swedish Species Information Centre, Swedish University of Agricultural Sciences, Box 7007, 12 SE-750 07 Uppsala, Sweden 13 c Percy FitzPatrick Institute of African Ornithology, DST/NRF Centre of Excellence, 14 University of Cape Town, Rondebosch 7700, South Africa 15 d Systematics and Biodiversity, Gothenburg University, Department of Zoology, Box 463, SE- 16 405 30 Göteborg, Sweden 17 e Bell Museum of Natural History and Department of Ecology, Evolution and Behavior, 18 University of Minnesota, 1987 Upper Buford Circle, St. Paul, MN 55108, USA 19 f Percy FitzPatrick Institute Centre of Excellence, Department of Genetics, University of 20 Pretoria, Hatfield, 0083, South Africa 21 g Institute of Pure & Applied Biology, Bahauddin Zakariya University, 60800, Multan, 22 Pakistan 23 h Department of Biology, Trent University, DNA Building, Peterborough, ON K9J 7B8, 24 Canada 25 i CEFE/CNRS Campus du CNRS 1919, route de Mende, 34293 Montpellier, France 26 27 * Corresponding author: Key Laboratory of Zoological Systematics and Evolution, Institute of 28 Zoology, Chinese Academy of Sciences, Chaoyang District, Beijing, 100101, P. R. China; E- 29 mail: [email protected] 30 1 31 ABSTRACT 32 The Alaudidae (larks) is a large family of songbirds in the superfamily Sylvioidea. -

Phenotypic Variation of Larks Along an Aridity Gradient Tieleman, BI; Williams, JB; Buschur, ME; Brown, CR
University of Groningen Phenotypic variation of larks along an aridity gradient Tieleman, BI; Williams, JB; Buschur, ME; Brown, CR Published in: Ecology DOI: 10.1890/0012-9658%282003%29084%5B1800%3APVOLAA%5D2.0.CO%3B2 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2003 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Tieleman, BI., Williams, JB., Buschur, ME., & Brown, CR. (2003). Phenotypic variation of larks along an aridity gradient: Are desert birds more flexible? Ecology, 84(7), 1800-1815. https://doi.org/10.1890/0012- 9658%282003%29084%5B1800%3APVOLAA%5D2.0.CO%3B2 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). The publication may also be distributed here under the terms of Article 25fa of the Dutch Copyright Act, indicated by the “Taverne” license. More information can be found on the University of Groningen website: https://www.rug.nl/library/open-access/self-archiving-pure/taverne- amendment. Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. -

EURASIAN SKYLARK Alauda Arvensis
EURASIAN SKYLARK Alauda arvensis Other: Sky Lark (1995-2015), A.a. arvensis/japonica? (naturalized) Common Skylark A.a. pekinensis (vagrant) naturalized (non-native) resident, long established; non-breeding visitor, vagrant The Eurasian Skylark breeds in temperate latitudes from W Europe and N Africa to Siberia and Japan, withdrawing S in winter to Africa, India, and Burma (Dement'ev and Gladkov 1954b, Ali and Ripley 1987, Cramp and Simmons 1988, Campbell et al. 1997, AOU 1998). It may have expanded its range eastward across these continents during the past millennia, aided by increased habitat afforded by human agricultural practices (Long 1981). It has been successfully introduced to Vancouver I, Australia, New Zealand, and a few other Pacific islands (Long 1981, Lever 1987, Campbell et al. 1997, Higgins et al. 2006). Eurasian Skylarks from sedentary European or Asian populations (see below) were introduced to the Southeastern Hawaiian Islands in 1865- 1870, and they continue to thrive on most islands, but since have disappeared or become scarce on Kaua'i and O'ahu, perhaps in part due to the conversion of ranch lands to agricultural fields (Munro 1944). They were introduced primarily for aesthetic purposes but there was some concern about their consuming newly planted crop seeds (Fisher 1948c; E 17:58, 17:81, 35:73; see Long 1981). In addition, Eurasian Skylarks from migratory ne. Asian populations have occurred and bred once in Alaska (AOU 1998), with single vagrants each reaching California (CBRC 2007) and Johnston Atoll (reported Nov 1963; Amerson and Shelton 1976) and two vagrants reaching the Northwestern Hawaiian Islands. -
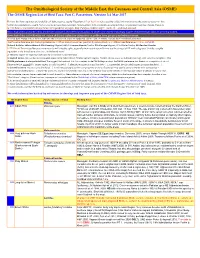
OSME List V3.4 Passerines-2
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa: Part C, Passerines. Version 3.4 Mar 2017 For taxa that have unproven and probably unlikely presence, see the Hypothetical List. Red font indicates either added information since the previous version or that further documentation is sought. Not all synonyms have been examined. Serial numbers (SN) are merely an administrative conveninence and may change. Please do not cite them as row numbers in any formal correspondence or papers. Key: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. The Passerine Reference List (including References for Hypothetical passerines [see Part E] and explanations of Abbreviated References) follows at Part D. Notes↓ & Status abbreviations→ BM=Breeding Migrant, SB/SV=Summer Breeder/Visitor, PM=Passage Migrant, WV=Winter Visitor, RB=Resident Breeder 1. PT=Parent Taxon (used because many records will antedate splits, especially from recent research) – we use the concept of PT with a degree of latitude, roughly equivalent to the formal term sensu lato , ‘in the broad sense’. 2. The term 'report' or ‘reported’ indicates the occurrence is unconfirmed. -

Systematic Notes on Asian Birds. 12.1 Types of the Alaudidae
ZV-335 085-126 | 12 03-01-2007 09:10 Pagina 85 Systematic notes on Asian birds. 12.1 Types of the Alaudidae E.C. Dickinson, R.W.R.J. Dekker, S. Eck & S. Somadikarta With contributions by M. Kalyakin, V. Loskot, H. Morioka, C. Violani, C. Voisin & J-F. Voisin Dickinson, E.C., R.W.R.J. Dekker, S. Eck & S. Somadikarta. Systematic notes on Asian birds. 12. Types of the Alaudidae. Zool. Verh. Leiden 335, 10.xii.2001: 85-126.— ISSN 0024-1652. Edward C. Dickinson, c/o The Trust for Oriental Ornithology, Flat 3, Bolsover Court, 19 Bolsover Road, Eastbourne, East Sussex, BN20 7JG, U.K. (e-mail: [email protected]). René W.R.J. Dekker, National Museum of Natural History, P.O. Box 9517, 2300 RA Leiden, The Netherlands. (e-mail: [email protected]). Siegfried Eck, Staatliche Naturhistorische Sammlungen Dresden, Museum für Tierkunde, A.B. Meyer Bau, Königsbrücker Landstrasse 159, D-01109 Dresden, Germany. (e-mail: [email protected]). Soekarja Somadikarta, Dept. of Biology, Faculty of Science and Mathematics, University of Indonesia, Depok Campus, Depok 16424, Indonesia. (e-mail: [email protected]). Mikhail V. Kalyakin, Zoological Museum, Moscow State University, Bol’shaya Nikitskaya Str. 6, Moscow 103009, Russia. (e-mail: [email protected]). Vladimir M. Loskot, Department of Ornithology, Zoological Institute, Russian Academy of Science, St. Petersburg, 199034, Russia. (e-mail: [email protected]). Hiroyuki Morioka, Curator Emeritus, National Science Museum, Hyakunin-cho 3-23-1, Shinjuku-ku, Tokyo 100, Japan. Carlo Violani, Department of Biology, University of Pavia, Piazza Botta 9, 27100 Pavia, Italy. -

See the Checklist
Official Reader Rendezvous Checklist Portugal: Birding an Ancient Land Lisbon, Portugal April 2017 ü (Common) Shelduck Tadorna tadorna ü Great Bustard Otis tarda ü Mallard Anas platyrhynchos ü Little Bustard Tetrax tetrax ü Gadwall Anas strepera ü (Eurasian) Oystercatcher Haematopus ostralegus ü (Northern) Shoveler Anas clypeata ü (Pied) Avocet Recurvirostra avosetta ü Garganey Anas querquedula ü Black-winged Stilt Himantopus himantopus ü (Common) Pochard Aythya ferina ü Stone Curlew Burhinus oedicnemus ü Red-crested Pochard Netta rufina ü Collared Pratincole Glareola pratincola ü Red-legged Partridge Alectoris rufa ü Little Ringed Plover Charadrius dubius ü (Common) Quail Coturnix coturnix ü (Common) Ringed Plover Charadrius hiaticula ü Little Grebe Tachybaptus ruficollis ü Kentish Plover Charadrius alexandrinus ü Great Crested Grebe Podiceps cristatus ü Grey Plover Pluvialis squatarola ü Cory's Shearwater Calonectris diomedea borealis ü (Northern) Lapwing Vanellus vanellus ü Balearic Shearwater Puffinus mauretanicus ü (Red) Knot Calidris canutus ü (Northern) Gannet Morus bassanus ü Sanderling Calidris alba ü (Great) Cormorant Phalacrocorax carbo ü (Ruddy) Turnstone Arenaria interpres ü (European) Shag Phalacrocorax aristotelis ü Dunlin Calidris alpina ü Little Bittern Ixobrychus minutus ü Curlew Sandpiper Calidris ferruginea ü (Black-crowned) Night Heron Nycticorax nycticorax ü Little Stint Calidris minuta ü Cattle Egret Bubulcus ibis ü Wood Sandpiper Tringa glareola ü Squacco Heron Ardeola ralloides ü Common Sandpiper Actitis hypoleucos -

Studies of Less Familiar Birds 116. Crested Lark by I
Studies of less familiar birds 116. Crested Lark By I. J. Ferguson-Lees Photographs by lb Trap-Lind (Plates 6-7) WE HAVE ONLY two photographs of the Crested Lark (Galerida cristatd) here, but they show the salient features very well. The species gets its name from the long upstanding crest which arises from the middle of its crown and which is conspicuous even when depressed. Many other larks have crests and inexperienced observers are sometimes misled by that on the Skylark (Alauda arvensis) when they see it raised and at close range, but the crest of the Crested Lark is really of almost comical proportions. The two plates also illustrate several other characters of this species and attention is drawn to these in the caption on plate 7. Crested Larks have a somewhat undulating flight, rather like that of Woodlarks (Lislbfa arborea), and a characteris tic outline from their short tails and broad, rounded wings. In Britain the Crested Lark is a surprisingly rare vagrant which has been recorded on less than fifteen occasions. The last two accepted observations were on Fair Isle in 1952 (Brit. Birds, 46: 211) and in Devonin 1958-5C) (Brit. Birds, 53: 167, 422), though it should be added that the species has almost certainly also occurred in Kent on at least two occasions in the last five years. Thus, while so many other birds formerly regarded as very rare wanderers to Britain are now known to be of annual occurrence in small numbers—the Melodious Warbler (Hippolais polyglottd) is one such example—the enormous increase in experienced observers has failed to raise the number of records of the Crested Lark. -
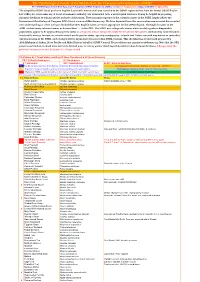
Simplified-ORL-2019-5.1-Final.Pdf
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa, Part F: Simplified OSME Region List (SORL) version 5.1 August 2019. (Aligns with ORL 5.1 July 2019) The simplified OSME list of preferred English & scientific names of all taxa recorded in the OSME region derives from the formal OSME Region List (ORL); see www.osme.org. It is not a taxonomic authority, but is intended to be a useful quick reference. It may be helpful in preparing informal checklists or writing articles on birds of the region. The taxonomic sequence & the scientific names in the SORL largely follow the International Ornithological Congress (IOC) List at www.worldbirdnames.org. We have departed from this source when new research has revealed new understanding or when we have decided that other English names are more appropriate for the OSME Region. The English names in the SORL include many informal names as denoted thus '…' in the ORL. The SORL uses subspecific names where useful; eg where diagnosable populations appear to be approaching species status or are species whose subspecies might be elevated to full species (indicated by round brackets in scientific names); for now, we remain neutral on the precise status - species or subspecies - of such taxa. Future research may amend or contradict our presentation of the SORL; such changes will be incorporated in succeeding SORL versions. This checklist was devised and prepared by AbdulRahman al Sirhan, Steve Preddy and Mike Blair on behalf of OSME Council. Please address any queries to [email protected].