Relating Individual Characteristics to Resting State Fmri Networks John Cocjin (Cocjinjb)1 , Jonathan Fisher (Jofisher)1 , Charles Chen2 1 2 CS224W, CS229 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Exploring the Sensorimotor Network Using Functional Connectivity and Graph Theory
EXPLORING THE SENSORIMOTOR NETWORK USING FUNCTIONAL CONNECTIVITY AND GRAPH THEORY by Ronald Bishop Submitted in partial fulfillment of the requirements for the degree of Master of Science at Dalhousie University Halifax, Nova Scotia August 2014 © Copyright by Ronald Bishop, 2014 TABLE OF CONTENTS LIST OF TABLES ......................................................................................................................................... v LIST OF FIGURES ...................................................................................................................................... vi ABSTRACT ................................................................................................................................................. vii LIST OF ABBREVIATIONS USED ......................................................................................................... viii ACKNOWLEDGEMENTS ......................................................................................................................... xi CHAPTER 1: INTRODUCTION ................................................................................................................. 1 CHAPTER 2: LITERATURE REVIEW ..................................................................................................... 4 2.1 AN OVERVIEW OF NEURONAL COMMUNICATION ..................................................................................... 4 2.2 SYNCHRONOUS NEURAL FIRING .............................................................................................................. -
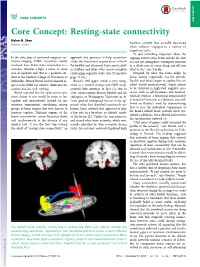
Core Concept: Resting-State Connectivity Helen H
CORE CONCEPTS CORE CONCEPTS Core Concept: Resting-state connectivity Helen H. Shen baseline activity that actually decreased Science Writer when subjects engaged in a variety of cognitive tasks. “It said something important about the In the early days of functional magnetic res- approach that promises to help researchers ongoing activity of the brain, and the fact that onance imaging (fMRI), researchers mostly study the functional organization of both it is not just sitting there waiting for someone analyzed how brain areas responded to a the healthy and abnormal brain, particularly in a white coat to come along and tell you stimulus, whether a light, a noise, or some in children and others who cannot complete what to do,” says Raichle. sort of cognitive task. But as a graduate stu- challenging cognitive tasks. (See Perspective Intrigued by what the brain might be dent at the Medical College of Wisconsin in page 14105.) doing during supposedly inactive periods, Milwaukee, Bharat Biswal had an unusual re- Biswal’s 1995 paper, which is now recog- Raichle and others began to explore this so- quest of his fMRI test subjects: climb into the nized as a seminal resting-state fMRI study, called “default mode network,” which seemed scanner and do, well, nothing. received little attention at first (1). But in to be involved in high-level cognitive pro- Biswal expected that the spontaneous neu- 2001, neuroscientist Marcus Raichle and his cesses, such as self-awareness and memory. ronal chatter at rest would be more or less colleagues at Washington University in St. Michael Greicius, a behavioral neuroscientist random and unstructured. -

Neuroimage: Clinical 15 (2017) 513–524
NeuroImage: Clinical 15 (2017) 513–524 Contents lists available at ScienceDirect NeuroImage: Clinical journal homepage: www.elsevier.com/locate/ynicl Disruption to control network function correlates with altered dynamic MARK connectivity in the wider autism spectrum ⁎ N. de Lacya,b, D. Dohertyb,c, B.H. Kingd, S. Rachakondae, V.D. Calhoune,f, a Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA 98195, USA b Seattle Children's Research Institute, Center for Integrative Brain Research, Seattle, WA 98105, USA c Department of Pediatrics, Divisions of Developmental and Genetic Medicine, University of Washington, Seattle, WA 98195, USA d Department of Psychiatry, University of California San Francisco, San Francisco, CA 94143, USA e The Mind Research Network & LBERI, Albuquerque, NM 87106, USA f Department of Electrical and Computer Engineering, University of New Mexico, Albuquerque, NM 87131, USA ARTICLE INFO ABSTRACT Keywords: Autism is a common developmental condition with a wide, variable range of co-occurring neuropsychiatric Autism symptoms. Contrasting with most extant studies, we explored whole-brain functional organization at multiple Functional MRI levels simultaneously in a large subject group reflecting autism's clinical diversity, and present the first network- Dynamic connectivity based analysis of transient brain states, or dynamic connectivity, in autism. Disruption to inter-network and inter- Control networks system connectivity, rather than within individual networks, predominated. We identified coupling disruption in Task-positive the anterior-posterior default mode axis, and among specific control networks specialized for task start cues and Default-mode the maintenance of domain-independent task positive status, specifically between the right fronto-parietal and cingulo-opercular networks and default mode network subsystems. -

Development of Thalamocortical Connectivity During Infancy and Its Cognitive Correlations
The Journal of Neuroscience, July 2, 2014 • 34(27):9067–9075 • 9067 Development/Plasticity/Repair Development of Thalamocortical Connectivity during Infancy and Its Cognitive Correlations Sarael Alcauter,1 Weili Lin,1 X J. Keith Smith,2 Sarah J. Short,3 Barbara D. Goldman,4 J. Steven Reznick,4 John H. Gilmore,3 and Wei Gao1 1Department of Radiology and Biomedical Research Imaging Center, 2Department of Radiology, 3Department of Psychiatry, and 4Frank Porter Graham Child Development Institute and Department of Psychology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599 Although commonly viewed as a sensory information relay center, the thalamus has been increasingly recognized as an essential node in various higher-order cognitive circuits, and the underlying thalamocortical interaction mechanism has attracted increasing scientific interest. However, the development of thalamocortical connections and how such development relates to cognitive processes during the earliest stages of life remain largely unknown. Leveraging a large human pediatric sample (N ϭ 143) with longitudinal resting-state fMRI scans and cognitive data collected during the first 2 years of life, we aimed to characterize the age-dependent development of thalamo- cortical connectivity patterns by examining the functional relationship between the thalamus and nine cortical functional networks and determine the correlation between thalamocortical connectivity and cognitive performance at ages 1 and 2 years. Our results revealed that the thalamus–sensorimotor -

Opposite Effects of Dopamine and Serotonin on Resting-State Networks: Review and Implications for Psychiatric Disorders
Molecular Psychiatry https://doi.org/10.1038/s41380-019-0406-4 REVIEW ARTICLE Opposite effects of dopamine and serotonin on resting-state networks: review and implications for psychiatric disorders 1,2 3 1,2,4,5 1,2 2,6,7 Benedetta Conio ● Matteo Martino ● Paola Magioncalda ● Andrea Escelsior ● Matilde Inglese ● 1,2 8,9,10 Mario Amore ● Georg Northoff Received: 15 September 2018 / Revised: 18 January 2019 / Accepted: 5 March 2019 © Springer Nature Limited 2019 Abstract Alterations in brain intrinsic activity—as organized in resting-state networks (RSNs) such as sensorimotor network (SMN), salience network (SN), and default-mode network (DMN)—and in neurotransmitters signaling—such as dopamine (DA) and serotonin (5-HT)—have been independently detected in psychiatric disorders like bipolar disorder and schizophrenia. Thus, the aim of this work was to investigate the relationship between such neurotransmitters and RSNs in healthy, by reviewing the relevant work on this topic and performing complementary analyses, in order to better understand their physiological link, as well as their alterations in psychiatric disorders. According to the reviewed data, neurotransmitters nuclei diffusively project to 1234567890();,: 1234567890();,: subcortical and cortical regions of RSNs. In particular, the dopaminergic substantia nigra (SNc)-related nigrostriatal pathway is structurally and functionally connected with core regions of the SMN, whereas the ventral tegmental area (VTA)-related mesocorticolimbic pathway with core regions of the SN. The serotonergic raphe nuclei (RNi) connections involve regions of the SMN and DMN. Coherently, changes in neurotransmitters activity impact the functional configuration and level of activity of RSNs, as measured by functional connectivity (FC) and amplitude of low-frequency fluctuations/temporal variability of BOLD signal. -

Human Cortical Sensorimotor Network Underlying Feedback Control of Vocal Pitch
Human cortical sensorimotor network underlying feedback control of vocal pitch Edward F. Changa,1,2, Caroline A. Niziolekb,1, Robert T. Knighta, Srikantan S. Nagarajanc, and John F. Houdeb,2 Departments of aNeurological Surgery, bOtolaryngology, and cRadiology, University of California, San Francisco, CA 94143 Edited* by Michael Merzenich, Brain Plasticity Institute, San Francisco, CA, and approved December 21, 2012 (received for review September 28, 2012) The control of vocalization is critically dependent on auditory feed- extracranial field potentials. ECoG monitoring has the advan- back. Here, we determined the human peri-Sylvian speech network tage of simultaneous high spatial and temporal resolution as well that mediates feedback control of pitch using direct cortical recordings. as the excellent signal-to-noise properties needed for single-trial Subjects phonated while a real-time signal processor briefly perturbed analyses. During neural recording, we used a digital signal-pro- their output pitch (speak condition). Subjects later heard the same cessing device (DSP) to induce real-time pitch perturbations recordings of their auditory feedback (listen condition). In posterior while subjects vocalized a prolonged vowel /ɑ/ sound (Fig. 1). superior temporal gyrus, a proportion of sites had suppressed re- The subject’s microphone signal was manipulated to create 200- sponses to normal feedback, whereas other spatially independent sites cent (two-semitone) upward or downward shifts in pitch (F0) and had enhanced responses to altered feedback. Behaviorally, speakers was fed back to the subject’s earphones (speak condition). This compensated for perturbations by changing their pitch. Single-trial pitch-shifted audio feedback was recorded and later played back analyses revealed that compensatory vocal changes were predicted by to subjects (listen condition). -

Impaired Effective Functional Connectivity of the Sensorimotor Network in Interictal Episodic Migraineurs Without Aura
Impaired Effective Functional Connectivity of the Sensorimotor Network in Interictal Episodic Migraineurs Without Aura Heng-Le Wei Nanjing Jiangning Hospital Jing Chen Nanjing Jiangning Hospital Yu-Chen Chen Nanjing First Hospital Yu-Sheng Yu Nanjing Jiangning Hospital Xi Guo Nanjing Jiangning Hospital Gang-Ping Zhou Nanjing Jiangning Hospital Qing-Qing Zhou Nanjing Jiangning Hospital Zhen-Zhen He Nanjing Jiangning Hospital Lian Yang Nanjing Jiangning Hospital Xindao Yin Nanjing First Hospital Junrong Li Nanjing Jiangning Hospital Hong Zhang ( [email protected] ) Jiangning Hospital Research article Keywords: magnetic resonance imaging, migraine, sensorimotor network, effective functional connectivity Posted Date: July 2nd, 2020 Page 1/19 DOI: https://doi.org/10.21203/rs.3.rs-39331/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on September 14th, 2020. See the published version at https://doi.org/10.1186/s10194-020-01176-5. Page 2/19 Abstract Background: Resting-state functional magnetic resonance imaging (Rs-fMRI) has conrmed sensorimotor network (SMN) dysfunction in migraine without aura (MwoA). However, the underlying mechanisms of SMN causal functional connectivity in MwoA remain unclear. We aimed to explore the association between clinical characteristics and effective functional connectivity in SMN, in interictal patients who have MwoA. Methods: We used Rs-fMRI to acquire imaging data in forty episodic patients with MwoA in the interictal phase and thirty-four healthy controls (HCs). Independent component analysis was used to prole the distribution of SMN and calculate the different SMN activity between the two groups. -

Activation of Brain Sensorimotor Network by Somatosensory Input in Patients with Hemiparetic Stroke: a Functional MRI Study
Chapter 4 Activation of Brain Sensorimotor Network by Somatosensory Input in Patients with Hemiparetic Stroke: A Functional MRI Study Hiroyuki Kato and Masahiro Izumiyama Additional information is available at the end of the chapter http://dx.doi.org/10.5772/51693 1. Introduction Stroke is one of the leading causes of disability in the elderly in many countries. Residual motor impairment, especially hemiparesis, is one of the most common sequelae after stroke. Motor recovery after stroke exhibits a wide range of difference among patients, and is dependent on the location and amount of brain damage, degree of impairment, and nature of deficit (Duncan et al., 1992). Full recovery of motor function is often observed when initial impairment is mild, but recovery is limited when there were severe deficits at stroke onset. The motor recovery after stroke may be caused by the effects of medical therapy against acute stroke, producing a resolution of brain edema and an increase in cerebral blood flow in the penumbra and remote areas displaying diaschisis. However, functional improvements may be seen past the period of acute tissue response and its resolution. The role of rehabilitation in facilitating motor recovery is considered to be produced by promoting brain plasticity. Non-invasive neuroimaging techniques, including functional magnetic resonance imaging (fMRI) and positron emission tomography (PET), enable us to measure task-related brain activity with excellent spatial resolution (Herholz & Heiss, 2000; Calautti & Baron, 2003; Rossini et al., 2003). The functional neuroimaging studies usually employ active motor tasks, such as hand grip and finger tapping, and require that the patients are able to move their hand. -

Cerebral Functional Networks During Sleep in Young and Older Individuals
www.nature.com/scientificreports OPEN Cerebral functional networks during sleep in young and older individuals Véronique Daneault1,2,3,12, Pierre Orban1,4,11, Nicolas Martin1,2,3, Christian Dansereau1, Jonathan Godbout2,5, Philippe Pouliot6, Philip Dickinson1, Nadia Gosselin2,3, Gilles Vandewalle7, Pierre Maquet7, Jean‑Marc Lina5,8,9,10, Julien Doyon1, Pierre Bellec1,12 & Julie Carrier1,2,3,12* Even though sleep modifcation is a hallmark of the aging process, age‑related changes in functional connectivity using functional Magnetic Resonance Imaging (fMRI) during sleep, remain unknown. Here, we combined electroencephalography and fMRI to examine functional connectivity diferences between wakefulness and light sleep stages (N1 and N2 stages) in 16 young (23.1 ± 3.3y; 7 women), and 14 older individuals (59.6 ± 5.7y; 8 women). Results revealed extended, distributed (inter‑ between) and local (intra‑within) decreases in network connectivity during sleep both in young and older individuals. However, compared to the young participants, older individuals showed lower decreases in connectivity or even increases in connectivity between thalamus/basal ganglia and several cerebral regions as well as between frontal regions of various networks. These fndings refect a reduced ability of the older brain to disconnect during sleep that may impede optimal disengagement for loss of responsiveness, enhanced lighter and fragmented sleep, and contribute to age efects on sleep‑dependent brain plasticity. Almost a third of human life is spent sleeping. Tis fundamental physiological state of reduced consciousness is composed of a series of alternating cycles, non-rapid eye movement (NREM) and rapid eye movement (REM) sleep. NREM state further decomposes into two light sleep stages (N1 and N2) and then to a deeper sleep (N3) stage. -

The Human Connectome Project Connectome Dysfunctions in PD
Connectome Dysfunction in Parkinson’s Disease ADEMOLA A. OREMOSU MBBS, MSc, PhD. International Parkinson and Movement Disorder Society | 555 East Wells Street, Suite 1100, Milwaukee WI 53202-3823 USA Tel: +1 414- 276-2145 | www.movementdisorders.org | [email protected] Objectives .Define Connectomes .Describe the basic connnections of the Brain and Connectoctomics .Describe the disorders of Connectomes in Parkinson’s Disease Outline 3 • Parkinson’s disease: What is it? Brief History Clinical and Neuropathological features • Synaptic and structural plasticity changes in PD Brain connectome and connectomics The Human Connectome Project Connectome dysfunctions in PD • Summary/conclusions 4 What is Parkinson’s disease (PD)? 5 •Disorder of the nervous system that primarily affects movements and bodily coordination. Brief History of PD 6 • In 175 AD, Galen described a condition characterised by tremors as ‘Shaking Palsy’. • Modern understanding of PD is attributed to James Parkinson and Jean-Martin Charcot. Types of PD 7 Global epidemiology of PD 8 • The most common movement disorder affecting 1 - 2 % of the general population over the age of 65 years. • The second most common neurodegenerative disorder after Dorsey, E. R., Elbaz, A., Nichols, E., Abd-Allah, F., Abdelalim, A., Adsuar, J. C. & Dahodwala, N. (2018). Global, regional, and national burden of Parkinson's disease, 1990–2016: a systematic analysis for the Alzheimer’s disease (AD) Global Burden of Disease Study 2016. The Lancet Neurology, 17(11), 939-953. Epidemiology of PD in Africa 9 Oluwole, O. G., Kuivaniemi, H., Carr, J. A., Ross, O. A., Olaogun, M. O., Bardien, S., & Komolafe, M. A. (2019). Parkinson's disease in Nigeria: A review of published studies and recommendations for future research. -

Large-Scale Brain Networks in Cognition: Emerging Methods and Principles
Review Feature Review Large-scale brain networks in cognition: emerging methods and principles Steven L. Bressler1 and Vinod Menon2 1 Center for Complex Systems and Brain Sciences, Department of Psychology, Florida Atlantic University, Boca Raton, FL, USA 2 Department of Psychiatry and Behavioral Sciences, Department of Neurology and Neurological Sciences, and Program in Neuroscience, Stanford University Medical School, Stanford, CA, USA An understanding of how the human brain produces cognition by revealing how cognitive functions arise from cognition ultimately depends on knowledge of large- interactions within and between distributed brain sys- scale brain organization. Although it has long been tems. It focuses on technological and methodological assumed that cognitive functions are attributable to advances in the study of structural and functional brain the isolated operations of single brain areas, we demon- connectivity that are inspiring new conceptualizations of strate that the weight of evidence has now shifted in large-scale brain networks. Underlying this focus is the support of the view that cognition results from the view that structure–function relations are critical for gain- dynamic interactions of distributed brain areas operat- ing a deeper insight into the neural basis of cognition. We ing in large-scale networks. We review current research thus emphasize the structural and functional architectures on structural and functional brain organization, and of large-scale brain networks (Box 1). For this purpose, we argue that the emerging science of large-scale brain networks provides a coherent framework for under- standing of cognition. Critically, this framework allows Glossary a principled exploration of how cognitive functions Blood-oxygen-level-dependent (BOLD) signal: measure of metabolic activity in emerge from, and are constrained by, core structural the brain based on the difference between oxyhemoglobin and deoxyhemo- globin levels arising from changes in local blood flow. -

Local-To-Distant Development of the Cerebrocerebellar Sensorimotor Network in the Typically Developing Human Brain: a Functional and Diffusion MRI Study
Brain Structure and Function (2019) 224:1359–1375 https://doi.org/10.1007/s00429-018-01821-5 ORIGINAL ARTICLE Local-to-distant development of the cerebrocerebellar sensorimotor network in the typically developing human brain: a functional and diffusion MRI study Kaoru Amemiya1 · Tomoyo Morita1,2 · Daisuke N. Saito3,4,5 · Midori Ban6 · Koji Shimada3,4 · Yuko Okamoto3,7 · Hirotaka Kosaka3,4,8 · Hidehiko Okazawa3,4 · Minoru Asada1,2 · Eiichi Naito1,9 Received: 4 October 2018 / Accepted: 16 December 2018 / Published online: 7 February 2019 © The Author(s) 2019 Abstract Sensorimotor function is a fundamental brain function in humans, and the cerebrocerebellar circuit is essential to this func- tion. In this study, we demonstrate how the cerebrocerebellar circuit develops both functionally and anatomically from child- hood to adulthood in the typically developing human brain. We measured brain activity using functional magnetic resonance imaging while a total of 57 right-handed, blindfolded, healthy children (aged 8–11 years), adolescents (aged 12–15 years), and young adults (aged 18–23 years) (n = 19 per group) performed alternating extension–flexion movements of their right wrists in precise synchronization with 1-Hz audio tones. We also collected their diffusion MR images to examine the extent of fiber maturity in cerebrocerebellar afferent and efferent tracts by evaluating the anisotropy-sensitive index of hindrance modulated orientational anisotropy (HMOA). During the motor task, although the ipsilateral cerebellum and the contralateral primary sensorimotor cortices were consistently activated across all age groups, the functional connectivity between these two distant regions was stronger in adults than in children and adolescents, whereas connectivity within the local cerebellum was stronger in children and adolescents than in adults.