Hyperion Manual Release 0.9.8
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Opensource Software in Mac OS X V. Zhhuta
Foss Lviv 2013 191 - Linux VM з Wordpress на Azure під’єднано до SQL-бази в приватному центрі обробки даних. Як бачимо, бізнес Microsoft вже дуже сильно зав'язаний на Open Source! Далі в доповіді будуть розглянуті подробиці інтероперабельності платформ з Linux Server, Apache Hadoop, Java, PHP, Node.JS, MongoDb, і наостанок дізнаємося про цікаві Open Source-розробки Microsoft Research. OpenSource Software in Mac OS X V. Zhhuta UK2 LImIted t/a VPS.NET, [email protected] Max OS X stem from Unix: bSD. It contains a lot of things that are common for Unix systems. Kernel, filesystem and base unix utilities as well as it's own package managers. It's not a secret that Mac OS X has a bSD kernel Darwin. The raw Mac OS X won't provide you with all power of Unix but this could be easily fixed: install package manager. There are 3 package manager: MacPorts, Fink and Homebrew. To dive in OpenSource world of mac os x we would try to install lates version of bash, bash-completion and few other utilities. Where we should start? First of all you need to install on you system dev-tools: Xcode – native development tools that contain GCC and libraries. Next step: bring a GIU – X11 into your system. Starting from Mac OS 10.8 X11 is not included in base-installation and it's need to install Xquartz(http://xquartz.macosforge.org). Now it's time to look closely to package managers MacPorts Site: www.macports.org Latest MacPorts release: 2.1.3 Number of ports: 16740 MacPorts born inside Apple in 2002. -

About Basictex-2021
About BasicTeX-2021 Richard Koch January 2, 2021 1 Introduction Most TeX distributions for Mac OS X are based on TeX Live, the reference edition of TeX produced by TeX User Groups across the world. Among these is MacTeX, which installs the full TeX Live as well as front ends, Ghostscript, and other utilities | everything needed to use TeX on the Mac. To obtain it, go to http://tug.org/mactex. 2 Basic TeX BasicTeX (92 MB) is an installation package for Mac OS X based on TeX Live 2021. Unlike MacTeX, this package is deliberately small. Yet it contains all of the standard tools needed to write TeX documents, including TeX, LaTeX, pdfTeX, MetaFont, dvips, MetaPost, and XeTeX. It would be dangerous to construct a new distribution by going directly to CTAN or the Web and collecting useful style files, fonts and so forth. Such a distribution would run into support issues as the creators move on to other projects. Luckily, the TeX Live install script has its own notion of \installation packages" and collections of such packages to make \installation schemes." BasicTeX is constructed by running the TeX Live install script and choosing the \small" scheme. Thus it is a subset of the full TeX Live with exactly the TeX Live directory structure and configuration scripts. Moreover, BasicTeX contains tlmgr, the TeX Live Manager software introduced in TeX Live 2008, which can install additional packages over the network. So it will be easy for users to add missing packages if needed. Since it is important that the install package come directly from the standard TeX Live distribution, I'm going to explain exactly how I installed TeX to produce the install package. -

Webroot Secureanywhere® Business – DNS Protection Apache License 2.0 • Aws-Sdk-Net Copyright © Amazon.Com, Inc. Apache
Webroot SecureAnywhere® Business – DNS Protection Apache License 2.0 • aws-sdk-net Copyright © Amazon.com, Inc. Apache License Version 2.0, January 2004 TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION 1. Definitions. “License” shall mean the terms and conditions for use, reproduction, and distribution as defined by Sections 1 through 9 of this document. “Licensor” shall mean the copyright owner or entity authorized by the copyright owner that is granting the License. “Legal Entity” shall mean the union of the acting entity and all other entities that control, are controlled by, or are under common control with that entity. For the purposes of this definition, “control” means (i) the power, direct or indirect, to cause the direction or management of such entity, whether by contract or otherwise, or (ii) ownership of fifty percent (50%) or more of the outstanding shares, or (iii) beneficial ownership of such entity. “You” (or “Your”) shall mean an individual or Legal Entity exercising permissions granted by this License. “Source” form shall mean the preferred form for making modifications, including but not limited to software source code, documentation source, and configuration files. “Object” form shall mean any form resulting from mechanical transformation or translation of a Source form, including but not limited to compiled object code, generated documentation, and conversions to other media types. “Work” shall mean the work of authorship, whether in Source or Object form, made available under the License, as indicated by a copyright notice that is included in or attached to the work (an example is provided in the Appendix below). -
MAC OSX Tips
Table of Contents MAC OSX tips keys linux-like environment and FINK MacPorts tips Build from source VS use binaries Useful commands MAC OSX tips Overview Some tips and hints on MacOSX usage: tame your keyboard and go ahead with compilation. edit 1414069126 [MAC OSX tips] section 1-168 1/8 documentation:tools:macosx_tips Edit keys quick copy and paste with CMD touch + c and + v key pipe | : alt + Maj + l tilde ~: alt +n square brackets [ or ]: alt + Maj + ( or ) antislash \:alt + maj + : edit 1414069126 [keys] section 169-383 documentation:tools:macosx_tips Edit linux-like environment and FINK 2/8 What is Fink? Fink is a distribution of Unix Open Source software for Mac OS X and Darwin. It brings a wide range of free command-line and graphical software developed for Linux and similar operating systems to your Mac. Quick Start Download the installer disk image: Fink 0.9.0 Binary Installer Double-click ?Fink-0.9.0-XYZ-Installer.dmg? to mount the disk image, then double-click the ?Fink 0.9.0 XYZ Installer.pkg? package inside. Follow the instructions on screen. At the end of the installation, the pathsetup utility will be launched. You will be asked for permission before your shell's configuration files are edited. When the utility has finished, you are set to go! Using the fink tool The fink tool uses several suffix commands to work on packages from the source distribution. Some of them need at least one package name, but can handle several package names at once. You can specify just the package name (e.g. -
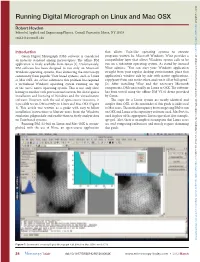
Running Digital Micrograph on Linux and Mac OSX
Downloaded from Running Digital Micrograph on Linux and Mac OSX https://www.cambridge.org/core Robert Hovden School of Applied and Engineering Physics, Cornell University, Ithaca, NY 14853 [email protected] Introduction that allows Unix-like operating systems to execute . IP address: Gatan Digital Micrograph (DM) software is considered programs written for Microsoft Windows. Wine provides a an industry standard among microscopists. The offline DM compatibility layer that allows Windows system calls to be run on a substitute operating system. As stated by internal application is freely available from Gatan [1]. Unfortunately, 170.106.202.58 DM software has been designed to run only on Microsoft Wine admins, “You can start your Windows application Windows operating systems, thus distancing the microscopy straight from your regular desktop environment, place that community from popular Unix-based systems, such as Linux application’s window side by side with native applications, or Mac OSX. An ad hoc solution to this problem has required copy/paste from one to the other, and run it all at full speed” , on a virtualized Windows operating system running on top [2]. After installing Wine and the necessary Microsoft 28 Sep 2021 at 14:01:56 of the user’s native operating system. This is not only slow, components, DM runs readily on Linux or OSX. The software having to emulate each processor instruction, but also requires has been tested using the offline DM V2.01 demo provided installation and licensing of Windows and the virtualization by Gatan. software. However, with the aid of open-source resources, it The steps for a Linux system are nearly identical and is possible to run DM natively on Linux and Mac OSX (Figure simpler than OSX, so the remainder of this guide is addressed , subject to the Cambridge Core terms of use, available at 1). -

Linux Package Management
Welcome A Basic Overview and Introduction to Linux Package Management By Stan Reichardt [email protected] October 2009 Disclaimer ● ...like a locomotive ● Many (similar but different) ● Fast moving ● Complex parts ● Another one coming any minute ● I have ridden locomotives ● I am NOT a locomotive engineer 2 Begin The Train Wreck 3 Definitions ● A file archiver is a computer program that combines a number of files together into one archive file, or a series of archive files, for easier transportation or storage. ● Metadata is data (or information) about other data (or information). 4 File Archivers Front Ends Base Package Tool CLI GUI tar .tar, tar tar file roller .tar.gz, .tgz, .tar.Z, .taz, .tar.bz2,.tbz2, .tbz, .tb2, .tar.lzma,.tlz, .tar.xz, .txz, .tz zip .zip zip zip file roller gzip gzip gunzip gunzip ● Archive file http://en.wikipedia.org/wiki/Archive_file ● Comparison of file archivers http://en.wikipedia.org/wiki/Comparison_of_file_archivers 5 tar ● These files end with .tar suffix. ● Compressed tar files end with “.t” variations: .tar.gz, .tgz, .tar.Z, .taz, .tar.bz2, .tbz2, .tbz, .tb2, .tar.lzma, .tlz, .tar.xz, .txz, .tz ● Originally intended for transferring files to and from tape, it is still used on disk-based storage to combine files before they are compressed. ● tar (file format) http://en.wikipedia.org/wiki/.tar 6 tarball ● A tar file or compressed tar file is commonly referred to as a tarball. ● The "tarball" format combines tar archives with a file-based compression scheme (usually gzip). ● Commonly used for source and binary distribution on Unix-like platforms, widely available elsewhere. -

1- Using Homebrew Package Manager 2- Using Macports Package Manager
This file describes how to install ATOMPAW on MacOS - Using Homebrew package manager - Using MacPorts package manager - Compiling by yourself This manpage is available as ~atompaw_src_dir/doc/README.MacOSX 1- USING HOMEBREW PACKAGE MANAGER A Homebrew third-party Formula for ATOMPAW is available Tested with macOS v10.9 to v10.14. Prerequisite: Homebrew installed (see: http://brew.sh/#install) To install ATOMPAW just type: brew install atompaw/repo/atompaw Note: libxc library is used by default. This can be disabled by passing --without-libxc option to brew install. 2- USING MACPORTS PACKAGE MANAGER ABINIT is available on MacPorts project, not necessarily in its latest version. Tested with mac OS v10.8 to 10.14. Prerequisites: § MacPorts installed (see: https://www.macports.org/install.php) § gcc (last version) port installed with Fortran variant (Fortran compiler), § Before starting, it is preferable to update MacPorts system: To install ABINIT just type: sudo port install atompaw By default, ATOMPAW is installed with the following libxc and accelerate (linear algebra) dependencies. Variant: Linking to OpenBLAS library: port install atompaw @X.Y.Z +openblas 3- COMPILING ATOMPAW BY YOURSELF under MacOSX Prerequisites: § MacOS (10.8+) § Xcode installed with command line tools (type: xcode-select --install) § A Fortran compiler installed. Possible options: - gfortran binary from: http://hpc.sourceforge.net - gfortran binary from: https://gcc.gnu.org/wiki/GFortranBinaries#MacOS - gfortran installed via a package manager (MacPorts, Homebrew, Fink) - Intel Fortran compiler § A Linear Algebra library. By default accelerate is included in MacOS. § By default the accelerate Framework is installed on MacOSX and ATOMPAW build system should find it. -

Pycg: Practical Call Graph Generation in Python
PyCG: Practical Call Graph Generation in Python Vitalis Salis,xy Thodoris Sotiropoulos,x Panos Louridas,x Diomidis Spinellisx and Dimitris Mitropoulosxz xAthens University of Economics and Business yNational Technical University of Athens zNational Infrastructures for Research and Technology - GRNET [email protected], ftheosotr, louridas, dds, [email protected] Abstract—Call graphs play an important role in different analysis. The primary aim for many implementations is com- contexts, such as profiling and vulnerability propagation analysis. pleteness, i.e., facts deduced by the system are indeed true [9]– Generating call graphs in an efficient manner can be a challeng- [11]. However, for dynamic languages, completeness comes ing task when it comes to high-level languages that are modular and incorporate dynamic features and higher-order functions. with a performance cost. Hence, such approaches are rarely Despite the language’s popularity, there have been very few employed in practice due to scalability issues [12]. This has tools aiming to generate call graphs for Python programs. Worse, led to the emergence of practical approaches focusing on in- these tools suffer from several effectiveness issues that limit their complete static analysis for achieving better performance [13], practicality in realistic programs. We propose a pragmatic, static [14]. Sacrificing completeness is the key enabler for adopt- approach for call graph generation in Python. We compute all assignment relations between program identifiers of functions, ing these approaches in applications that interact with com- variables, classes, and modules through an inter-procedural plex libraries [13], or Integrated Development Environments analysis. Based on these assignment relations, we produce the (IDEs) [14]. -

COMPUTING for SCIENCE and ENGINEERING Revised Through End of Chapter 6
COMPUTING FOR SCIENCE AND ENGINEERING Revised through end of Chapter 6 Robert D. Skeel This work is licensed under the Creative Commons Attribution 3.0 United StatesLicense. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/us/ or send a letter to Creative Commons, 444 Castro Street, Suite 900, Mountain View, California, 94041, USA. Contents 1 Introduction 1 1.1 Role of Computing in Science and Engineering.....................1 1.2 Using Unix.........................................2 1.2.1 Text editors.....................................5 1.2.2 Integrated development environments......................5 1.2.3 Shell scripts.....................................5 1.2.4 Unix tools......................................6 1.3 Installation of Software..................................8 1.3.1 Unix, C compiler, and OpenMP.........................9 1.3.2 The Anaconda Python Distribution and MPI.................. 10 1.3.3 ∗ The Enthought Python Distribution...................... 11 1.3.4 ∗ Separate Installation of Python and Packages................. 12 1.3.5 ∗ MPI for Python................................. 12 1.4 Use of Rosen Center Linux Cluster............................ 13 1.4.1 Remote access and file transfer.......................... 13 1.4.2 Python....................................... 13 2 Python 14 2.1 Interactive Computing................................... 14 2.1.1 Importing a module................................ 15 2.1.2 Documentation................................... 15 2.1.3 Lists........................................ -

VIP Documentation Release 0.8.1
VIP Documentation Release 0.8.1 Carlos Gomez Gonzalez, Olivier Wertz & VORTEX team Nov 17, 2017 Contents 1 Attribution 3 2 Introduction 5 3 Jupyter notebook tutorial 7 4 Mailing list 9 5 Quick Setup Guide 11 6 Installation and dependencies 13 6.1 Installation and dependencies...................................... 13 6.2 Loading VIP............................................... 14 7 Frequently asked questions 15 7.1 FAQ.................................................... 15 8 Package structure 17 8.1 vip package................................................ 19 9 API 21 i ii VIP Documentation, Release 0.8.1 Contents 1 VIP Documentation, Release 0.8.1 2 Contents CHAPTER 1 Attribution Please cite Gomez Gonzalez et al. 2016 (submitted) whenever you publish data reduced with VIP. Astrophysics Source Code Library reference [ascl:1603.003] 3 VIP Documentation, Release 0.8.1 4 Chapter 1. Attribution CHAPTER 2 Introduction VIP is an open source package/pipeline for angular, reference star and spectral differential imaging for exoplanet/disk detection through high-contrast imaging. VIP is being developed by the VORTEX team and collaborators. You can check on github the direct contributions to the code (tab contributors). Most of VIP‘s functionalities are mature but it doesn’t mean it’s free from bugs. The code will continue evolving and therefore all the feedback and contributions will be greatly appreciated. If you want to report a bug, suggest or add a functionality please create an issue or send a pull request on the github repository. The goal of VIP is to incorporate robust, efficient, easy-to-use, well-documented and open source implementations of high-contrast image processing algorithms to the interested scientific community. -

UA37/44 Tidbits of Kentucky Folklore
Western Kentucky University TopSCHOLAR® Faculty/Staff Personal Papers WKU Archives Records 1935 UA37/44 Tidbits of Kentucky Folklore Gordon Wilson Follow this and additional works at: https://digitalcommons.wku.edu/fac_staff_papers Part of the Folklore Commons, Linguistic Anthropology Commons, Oral History Commons, and the Social History Commons This Article is brought to you for free and open access by TopSCHOLAR®. It has been accepted for inclusion in Faculty/Staff Personal Papers by an authorized administrator of TopSCHOLAR®. For more information, please contact [email protected]. ORIGINALS TIDBITS OF KENTUCKY FOLKLORE by Gordon Wilson Vol. I Nos. 1-100 TIDBITS OF KENTUCKY FOLKLORE SERIES1 W. K, T. C. by Dr, Gordon Wilson Artfole I V Introduotion I/ .Artiole XXIV Passing Institutions- Dresses, Breeohes, and Artiole II V Folk Psyohology Pants, Part I, Article III ✓Breakdowns /Artiole XXV Passing Institutions- Dresses, Breeohes, and .Artiole IV V Play-Party Games Pants, Part II. .Artiole V V Kentucky Ballads and Songs Article XXVI t/Passing Institutions- Up On The Joist Article VI /The Virginia Reel v/ .Article XXVII Passing Institutions- Artiole VII V Fiddles and Violins The Home-l{ni t Yarn Stooking, Part I. Artiole VIII l,,1rild Greens Artiole XXVIII Passing Institutions- .Artiole IX V Folk Prejudioes The HOJ11e-Knit Yarn Stooking, Part II. Artiole X /'Passing Institutione-- ChipB Artiole XXIX Passing Institutions- The Home-Knit Yarn Article XI ✓Passing Institut1.ons- Stooking, Part IIIe Gourds· 1--/ - A....-ti <!;le X1I tt\5:s.i"''i rV\s-t'i11.. rb-i11~-- ~ / ,-rc.,it-ts V.btiole XXX Friday Afternoons .Artiole XIII VPassing Institutions-- The Old Fe.11\ily Nag, Pt. -

Portable Software-Installation Mit Pkgsrc
Universität Bielefeld Rainer Orth Portable Software-Installation mit pkgsrc Dipl.-Chem. Rainer Orth Technische Fakultät Universität Bielefeld [email protected] AG Rechnerbetrieb SS 2006 pkgsrc 1 von 12 Universität Bielefeld Rainer Orth Übersicht • Problemstellung • Benutzersicht • Parallelinstallationen: Package Views • Entwicklersicht • Probleme • Alternativen AG Rechnerbetrieb SS 2006 pkgsrc 2 von 12 Universität Bielefeld Rainer Orth Software-Installation: Der Status Quo • manuelles Bauen aus den Quellen für verschiedene Plattformen – zeitaufwendig – mühselig – verschiedene Versionsstände, keine problemlosen Updates wegen Abhängigkeiten – zunehmende Zahl von Abhängigkeiten – zahlreiche Packages kommen mit Volume-Struktur nicht gut zurecht – kein Tracking installierter Software AG Rechnerbetrieb SS 2006 pkgsrc 3 von 12 Universität Bielefeld Rainer Orth Problemstellung • Ziel: einheitliche plattformübergreifende Software-Installation • mindestens für Linux, Mac OS X, Solaris; gut, wenn auch IRIX und Tru64 UNIX • Option: native Package-Systeme (Blastwave, Sun Freeware, Fink, . ) – Einzelmaschinen-orientiert – plattformabhängig – keine einfache lokale Konfiguration – Binary Packages stellen verschiedene Software-Versionen in verschiedenen Konfigurationen bereit – schwierige Parallel-Installation verschiedener Versionen AG Rechnerbetrieb SS 2006 pkgsrc 4 von 12 Universität Bielefeld Rainer Orth pkgsrc • Framework zum Bauen und Installieren von Software-Paketen • entstanden aus FreeBSD ports, ursprünglich nur für NetBSD (August 1997)