2 Spitting Image: Decode Me!
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mtv and Transatlantic Cold War Music Videos
102 MTV AND TRANSATLANTIC COLD WAR MUSIC VIDEOS WILLIAM M. KNOBLAUCH INTRODUCTION In 1986 Music Television (MTV) premiered “Peace Sells”, the latest video from American metal band Megadeth. In many ways, “Peace Sells” was a standard pro- motional video, full of lip-synching and head-banging. Yet the “Peace Sells” video had political overtones. It featured footage of protestors and police in riot gear; at one point, the camera draws back to reveal a teenager watching “Peace Sells” on MTV. His father enters the room, grabs the remote and exclaims “What is this garbage you’re watching? I want to watch the news.” He changes the channel to footage of U.S. President Ronald Reagan at the 1986 nuclear arms control summit in Reykjavik, Iceland. The son, perturbed, turns to his father, replies “this is the news,” and lips the channel back. Megadeth’s song accelerates, and the video re- turns to riot footage. The song ends by repeatedly asking, “Peace sells, but who’s buying?” It was a prescient question during a 1980s in which Cold War militarism and the nuclear arms race escalated to dangerous new highs.1 In the 1980s, MTV elevated music videos to a new cultural prominence. Of course, most music videos were not political.2 Yet, as “Peace Sells” suggests, dur- ing the 1980s—the decade of Reagan’s “Star Wars” program, the Soviet war in Afghanistan, and a robust nuclear arms race—music videos had the potential to relect political concerns. MTV’s founders, however, were so culturally conserva- tive that many were initially wary of playing African American artists; addition- ally, record labels were hesitant to put their top artists onto this new, risky chan- 1 American President Ronald Reagan had increased peace-time deicit defense spending substantially. -

The Oxford Dictionary of New Words: a Popular Guide to Words in the News
The Oxford Dictionary of New Words: A popular guide to words in the news PREFACE Preface This is the first dictionary entirely devoted to new words and meanings to have been published by the Oxford University Press. It follows in the tradition of the Supplement to the Oxford English Dictionary in attempting to record the history of some recent additions to the language, but, unlike the Supplement, it is necessarily very selective in the words, phrases, and meanings whose stories it sets out to tell and it stands as an independent work, unrelated (except in the resources it draws upon) to the Oxford English Dictionary. The aim of the Oxford Dictionary of New Words is to provide an informative and readable guide to about two thousand high-profile words and phrases which have been in the news during the past decade; rather than simply defining these words (as dictionaries of new words have tended to do in the past), it also explains their derivation and the events which brought them to prominence, illustrated by examples of their use in journalism and fiction. In order to do this, it draws on the published and unpublished resources of the Oxford English Dictionary, the research that is routinely carried out in preparing new entries for that work, and the word-files and databases of the Oxford Dictionary Department. What is a new word? This, of course, is a question which can never be answered satisfactorily, any more than one can answer the question "How long is a piece of string?" It is a commonplace to point out that the language is a constantly changing resource, growing in some areas and shrinking in others from day to day. -
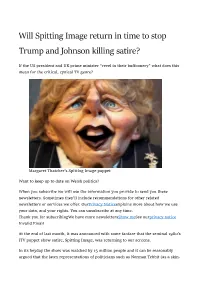
Will Spitting Image Return in Time to Stop Trump and Johnson Killing Satire?
Will Spitting Image return in time to stop Trump and Johnson killing satire? If the US president and UK prime minister “revel in their buffoonery” what does this mean for the critical, cynical TV genre? Margaret Thatcher's Spitting Image puppet Want to keep up to date on Welsh politics? When you subscribe we will use the information you provide to send you these newsletters. Sometimes they’ll include recommendations for other related newsletters or services we offer. OurPrivacy Noticeexplains more about how we use your data, and your rights. You can unsubscribe at any time. Thank you for subscribingWe have more newslettersShow meSee ourprivacy notice Invalid Email At the end of last month, it was announced with some fanfare that the seminal 1980’s ITV puppet show satire, Spitting Image, was returning to our screens. In its heyday the show was watched by 15 million people and it can be reasonably argued that the latex representations of politicians such as Norman Tebbit (as a skin- head bovver boy), Kenneth Baker (half human, half slug) and Michael Heseltine (a wide-eyed and feral Tarzan) filtered down into popular consciousness rendering the fictional far more memorable than the actual. Apparently, the proposed new series’ first episode has already been filmed and Roger Law, one of the show’s original creators, confirmed to the Guardian that producers were in talks with US TV networks about bringing the programme to a global audi- ence. This is significant because the primary target of the newest incarnation of Spitting Image is going to be, unsurprisingly, the so-called leader of the free world, Donald Trump. -

Television and the Cold War in the German Democratic Republic
0/-*/&4637&: *ODPMMBCPSBUJPOXJUI6OHMVFJU XFIBWFTFUVQBTVSWFZ POMZUFORVFTUJPOT UP MFBSONPSFBCPVUIPXPQFOBDDFTTFCPPLTBSFEJTDPWFSFEBOEVTFE 8FSFBMMZWBMVFZPVSQBSUJDJQBUJPOQMFBTFUBLFQBSU $-*$,)&3& "OFMFDUSPOJDWFSTJPOPGUIJTCPPLJTGSFFMZBWBJMBCMF UIBOLTUP UIFTVQQPSUPGMJCSBSJFTXPSLJOHXJUI,OPXMFEHF6OMBUDIFE ,6JTBDPMMBCPSBUJWFJOJUJBUJWFEFTJHOFEUPNBLFIJHIRVBMJUZ CPPLT0QFO"DDFTTGPSUIFQVCMJDHPPE Revised Pages Envisioning Socialism Revised Pages Revised Pages Envisioning Socialism Television and the Cold War in the German Democratic Republic Heather L. Gumbert The University of Michigan Press Ann Arbor Revised Pages Copyright © by Heather L. Gumbert 2014 All rights reserved This book may not be reproduced, in whole or in part, including illustrations, in any form (be- yond that copying permitted by Sections 107 and 108 of the U.S. Copyright Law and except by reviewers for the public press), without written permission from the publisher. Published in the United States of America by The University of Michigan Press Manufactured in the United States of America c Printed on acid- free paper 2017 2016 2015 2014 5 4 3 2 A CIP catalog record for this book is available from the British Library. ISBN 978– 0- 472– 11919– 6 (cloth : alk. paper) ISBN 978– 0- 472– 12002– 4 (e- book) Revised Pages For my parents Revised Pages Revised Pages Contents Acknowledgments ix Abbreviations xi Introduction 1 1 Cold War Signals: Television Technology in the GDR 14 2 Inventing Television Programming in the GDR 36 3 The Revolution Wasn’t Televised: Political Discipline Confronts Live Television in 1956 60 4 Mediating the Berlin Wall: Television in August 1961 81 5 Coercion and Consent in Television Broadcasting: The Consequences of August 1961 105 6 Reaching Consensus on Television 135 Conclusion 158 Notes 165 Bibliography 217 Index 231 Revised Pages Revised Pages Acknowledgments This work is the product of more years than I would like to admit. -

Its Only a Movie: Reel Life Adventures of a Film Obsessive Free
FREE ITS ONLY A MOVIE: REEL LIFE ADVENTURES OF A FILM OBSESSIVE PDF Mark Kermode | 352 pages | 01 Jan 2011 | Cornerstone | 9780099543480 | English | London, United Kingdom It's Only a Movie by Mark Kermode | Waterstones Goodreads helps you keep track of books you want to read. Want to Read saving…. Want to Read Currently Reading Read. Other editions. Enlarge cover. Error rating book. Refresh and try again. Open Preview See a Problem? Details if other :. Thanks for telling us about the problem. Return to Book Page. It's Only a Movie by Mark Kermode. Film critic and broadcaster, Mark Kermode, has written a forthright account of a life lived in film. From his favourite movies to rants against Pirates of the Caribbeanand the current resurgence of 3D, there are spirited defences of Hannah Montana and Mamma Mia. Get A Copy. Paperbackpages. More Details Original Title. Other Editions 6. Friend Reviews. To see what your friends thought of this book, please sign up. To ask other readers questions about It's Only a Movieplease sign up. Lists with This Book. Community Reviews. Showing Average rating 3. Rating details. More filters. Sort order. Start your review of It's Only a Movie. Especially for a book I bought two years ago to find info that wasn't even in it, this was a lot of fun. The first couple of chapters I liked so much I thought I'd be giving it 5 stars. The nostalgia was perfectly pitched - childhood 70s cinema, 80s leftwing student politics and journalism. That was being a "proper student", because of course, being a kid at the time, that's when my idea of how students were was formed. -

Antinuclear Politics, Atomic Culture, and Reagan Era Foreign Policy
Selling the Second Cold War: Antinuclear Cultural Activism and Reagan Era Foreign Policy A dissertation presented to the faculty of the College of Arts and Sciences of Ohio University In partial fulfillment of the requirements for the degree Doctor of Philosophy William M. Knoblauch March 2012 © 2012 William M. Knoblauch. All Rights Reserved. 2 This dissertation titled Selling the Second Cold War: Antinuclear Cultural Activism and Reagan Era Foreign Policy by WILLIAM M. KNOBLAUCH has been approved for the Department of History and the College of Arts and Sciences by __________________________________ Chester J. Pach Associate Professor of History __________________________________ Howard Dewald Dean, College of Arts and Sciences 3 ABSTRACT KNOBLAUCH, WILLIAM M., Ph.D., March 2012, History Selling the Second Cold War: Antinuclear Cultural Activism and Reagan Era Foreign Policy Director of Dissertation: Chester J. Pach This dissertation examines how 1980s antinuclear activists utilized popular culture to criticize the Reagan administration’s arms buildup. The 1970s and the era of détente marked a decade-long nadir for American antinuclear activism. Ronald Reagan’s rise to the presidency in 1981 helped to usher in the “Second Cold War,” a period of reignited Cold War animosities that rekindled atomic anxiety. As the arms race escalated, antinuclear activism surged. Alongside grassroots movements, such as the nuclear freeze campaign, a unique group of antinuclear activists—including publishers, authors, directors, musicians, scientists, and celebrities—challenged Reagan’s military buildup in American mass media and popular culture. These activists included Fate of the Earth author Jonathan Schell, Day After director Nicholas Meyer, and “nuclear winter” scientific-spokesperson Carl Sagan. -

Q27 Webs Review EN.Pdf 71.6 KB
Webs Review Latex parody United Kingdom Argentina Spitting Image Kanal K <http://www.museum.tv/archives/etv/S/htmlS/spittingimag/s <http://www.thefileroom.org/documents/dyn/DisplayCase.cf pittingimag.htm> m/id/47> Spitting Image was a satirical programme using rubber Until the beginning of the nineties, the channel Telefe puppets, broadcast on ITV from 1984 to 1996. This pro- broadcast the satirical programme Kanal K, starring rubber gramme created a genre that inspired other programmes puppets. In one programme, the puppet of Pope John Paul parodying political and social events in many other coun- II appeared, using gross language. At one time he used the tries. The arrival of Have I got news for you in 1990 brought Italian phrase “va fangullo” (“up yours”), which led to ener- strong competition that gradually displaced it. The pro- getic protests by the Argentinean Catholic church. Telefe gramme was cancelled at the start of 1996. stopped broadcasting the programme as a consequence of In spite of its criticism of politicians of the time (Margaret this controversy. Thatcher and Ronald Reagan), Spitting Image did not cause too much of a dilemma between decency and free speech. Australia However, BBC Radio refused to broadcast the programme's Rubbery Figures first hit song, with the puppet of Prince Andrew boasting, <http://www.nicholsoncartoons.com.au/rubbery.php> “I’m just a Prince who can’t say No”. Rubbery Figures was the series of political and social pa- rodies by the Australian TV channel ABC (Australian Broad- Germany casting Corporation). It was broadcast for six years (1984- Hurra Deutschland 1990), during which time it received a lot of criticism and <http://www.hurra-deutschland.de/> pressure, leading to production being stopped. -

WWII Question of “Why We !Ght” with “For the Soldiers Themselves.” Forrest Gump W!/ W" “S)00$%# #!" T%$$0.”: R!"#$%&'-+ E1$+)#&$2
W!" W# “S$%%&'( (!# T'&&%)”: R!#(&'*+,- E.&-$(*&/) R!"#$ S%&'( ) is essay tracks the genealogy of the contemporary call to “support the troops,” a rhetoric that includes but goes beyond the strategic and argumentative use of the phrase itself. Support-the-troops rhetoric has two major functions: de*ection and dissociation. De*ection involves discursive trends in play since Vietnam that have rede+ned war as a +ght to save our own soldiers—especially the captive soldier—rather than as a struggle for policy goals external to the military. As such, this discourse directs civic attention away from the question of whether the particular war policy is just. ) e essay explicates these trends through an examination of the POW/MIA, war +lm, and the symbol of the yellow ribbon. ) e second trope, dissociation, quarantines the citizen from questions of military action by manufacturing distance between citizen and soldier. Dissociation o, en goes further to de+ne civic deliberation and dissent as an attack on the soldier body and thus an ultimate immoral act. ) is essay explores this trope through executive rhetoric, an analysis of the particular phrase “support the troops,” metaphor for war, and John Kerry’s run for the presidency in -../. Both de*ection and dissociation work to discipline and mute public deliberation in matters of war. ) e essay concludes by considering strategies for reopening spaces for democratic deliberation. R!"#$ S%&'( is Assistant Professor of Speech Communication at the University of Georgia in Athens. © 2009 Michigan State University Board of Trustees. All rights reserved. Rhetoric & Public Affairs Vol. 12, No. 4, 2009, pp. -

The United States and the Vietnam War: a Guide to Materials at the British Library
THE BRITISH LIBRARY THE UNITED STATES AND THE VIETNAM WAR: A GUIDE TO MATERIALS AT THE BRITISH LIBRARY by Jean Kemble THE ECCLES CENTRE FOR AMERICAN STUDIES THE UNITED STATES AND THE VIETNAM WAR Introduction Bibliographies, Indexes, and other Reference Aids Background and the Decision to Intervene The Congressional Role The Executive Role General Roosevelt Truman Eisenhower Kennedy Johnson Nixon Ford Carter Constitutional and International Law The Media Public Opinion Anti-war Protests/Peace Activists Contemporary Analysis Retrospective Analysis Legacy: Domestic Legacy: Foreign Policy Legacy: Cultural Art Film and Television Novels, Short Stories and Drama Poetry Literary Criticism Legacy: Human Vietnamese Refugees and Immigrants POW/MIAs Oral Histories, Memoirs, Diaries, Letters Veterans after the War Introduction It would be difficult to overstate the impact on the United States of the war in Vietnam. Not only did it expose the limits of U.S. military power and destroy the consensus over post-World War II foreign policy, but it acted as a catalyst for enormous social, cultural and political upheavals that still resonate in American society today. This guide is intended as a bibliograhical tool for all those seeking an introduction to the vast literature that has been written on this subject. It covers the reasons behind American intervention in Vietnam, the role of Congress, the Executive and the media, the response of the American public, particularly students, to the escalation of the war, and the war’s legacy upon American politics, culture and foreign policy. It also addresses the experiences of those individuals affected directly by the war: Vietnam veterans and the Indochinese refugees. -

(CUWS) Outreach Journal #1166
USAF Center for Unconventional Weapons Studies (CUWS) Outreach Journal 22 May 2015 Feature Item: “Annual Report to Congress: Military and Security Developments involving the People’s Republic of China 2015”. Prepared by the U.S. Department of Defense; April 07, 2015; 98 pages. http://www.defense.gov/pubs/2015_China_Military_Power_Report.pdf The People’s Republic of China (PRC) continues to pursue a long-term, comprehensive military modernization program designed to improve its armed forces’ capacity to fight short-duration, high-intensity regional conflicts. Preparing for potential conflict in the Taiwan Strait remains the focus and primary driver of China’s military investment; however, the PRC is increasing its emphasis on preparations for contingencies other than Taiwan, such as contingencies in the East China Sea and South China Sea. Additionally, as China’s global footprint and international interests grow, its military modernization program has become progressively more focused on investments for a range of missions beyond China’s periphery, including power projection, sea lane security, counter-piracy, peacekeeping, and humanitarian assistance/disaster relief (HA/DR). U.S. Counter-WMD 1. Missile Defense Dispute Reignited 2. Countering Russian Strategic Missile Threat Too Hard, Costly for US 3. US Aegis Ships Could Pose Threat to Russia - Russian Diplomat U.S. Arms Control 1. US Defense Bill Bans Funding of Russian Nuclear Nonproliferation Efforts 2. Foreign Ministry: US Actions May Push Russia to Building up Nuclear Arsenals Homeland Security/The Americas 1. Air Force Getting Closer to Testing Hypersonic Weapon, Engineers Say Asia/Pacific 1. Kerry Hopes Iran Deal to Have 'Positive Influence' on N. -

Thesis Final
Historicising Neoliberal Britain: Remembering the End of History A thesis submitted to The University of Manchester for the degree of Doctor of Philosophy in the Faculty of Humanities 2018 Christopher R Vardy School of Arts, Languages and Cultures List of Contents Abstract 4 Declaration and Copyright Statement 5 Acknowledgements 6 Introduction – Remembering the End of History Historicising neoliberal Britain 8 ‘Maggie’, periodisation and collective memory of the 1980s 15 Thatcherism and Neoliberalism 23 The End of History 28 Historical fictions, historicisation and historicity 34 Thesis structure 37 Chapter One – The End of History Introduction: Origin Myth 40 Histories 44 Fictions 52 Historicising the End of History 59 Struggle and inevitability 66 A War of Ghosts / History from Below 72 Conclusion: Historicity without futurity 83 Chapter Two – No Future Introduction: ‘Our little systems have their day; They have their day and cease to be’ 86 Bodily permeabilities 94 The financialised imaginary 97 Shaping the 1980s: reprise and the ‘light of the moment’ 104 Cocaine economics 110 Embodied crises of futurity 115 Conclusion: Dissonance 121 Chapter Three – Thatcher’s Children: Neoliberal Adolescence Introduction: Genesis or Preface? 123 2 Retro-memory 126 ‘You’ve obviously forgotten what it’s like’ 132 The nuclear 1980s 137 A brutal childhood 142 Adolescence and critique 147 Conclusion: Perpetual Adolescence 156 Chapter Four – Thatcher’s Children: Abusive Historicity Introduction: ‘Lost Boys’ and the arrested bildungsroman 158 The historical child: uses and abuses 162 Precarious Futures: Death of a Murderer 171 Re-writing the James Bulger murder: ‘faultline narratives’ and The Field of Blood 175 Cycles of abuse: Nineteen Eighty Three 183 Conclusion: Abusive historicity 188 Conclusion – Dissonance and Critique 189 Works cited 195 Word count: 72,506 3 Abstract This thesis argues that a range of twenty-first-century British historical fictions historicise contemporary neoliberal politics, economics and subject-formation through a return to the Thatcherite past. -

Beating the Americans at Their Own Game an Offset Strategy with Chinese Characteristics
JUNE 2019 Beating the Americans at their Own Game An Offset Strategy with Chinese Characteristics Robert O. Work and Greg Grant About the Authors About the Defense Program ROBERT O. WORK is the Distinguished Over the past 10 years, CNAS has defined the future of Senior Fellow for Defense and National U.S. defense strategy. Building on this legacy, the CNAS Security at the Center for a New American Defense team continues to develop high-level concepts Security and the owner of TeamWork, and concrete recommendations to ensure U.S. military LLC, which specializes in national security preeminence into the future and to reverse the erosion of affairs and the future of warfare. Mr. Work U.S. military advantages vis-à-vis China, and to a lesser previously served as the Deputy Secretary extent Russia. Specific areas of study include concentrating of Defense, where he was responsible for overseeing the on great-power competition, developing a force structure day-to-day business of the Pentagon and developing the and innovative operational concepts adapted for this department’s $600 billion defense program. He is widely more challenging era, and making hard choices to effect credited for his work with leaders in the department and necessary change. the intelligence community on the “Third Offset Strategy,” which aimed to restore U.S. conventional overmatch over its strategic rivals and adversaries. He was awarded DoD’s Distinguished Public Service Award (twice), the National Intelligence Distinguished Public Service Award, and the Chairman of the Joint Chiefs of Staff Joint Distinguished Civilian Service Award. GREG GRANT is an Adjunct Senior Fellow for the Defense Program at the Center for a New American Security (CNAS).