Virus Discovery in All Three Major Lineages of Terrestrial Arthropods Highlights the Diversity of Single-Stranded DNA Viruses Associated with Invertebrates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Viral Infection in the Copepods Labidocera Aestiva And
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2012 Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida Darren Stephenson Dunlap University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Other Oceanography and Atmospheric Sciences and Meteorology Commons, and the Virology Commons Scholar Commons Citation Dunlap, Darren Stephenson, "Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida" (2012). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/4032 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Evidence of Viruses in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida By Darren S. Dunlap A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science College of Marine Science University of South Florida Major Professor: Mya Breitbart, Ph.D Kendra Daly, Ph.D Ian Hewson, Ph.D Date of Approval: March 19, 2012 Key Words: Copepods, Single-stranded DNA Viruses, Mesozooplankton, Transmission Electron Microscopy, Metagenomics Copyright © 2012, Darren Stephenson Dunlap DEDICATION None of this would have been possible without the generous love and support of my entire family over the years. My parents, Steve and Jill Dunlap, have always encouraged my pursuits with support and love, and their persistence of throwing me into lakes and rivers is largely responsible for my passion for Marine Science. -

Pdf Available
Virology 554 (2021) 89–96 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/virology Diverse cressdnaviruses and an anellovirus identifiedin the fecal samples of yellow-bellied marmots Anthony Khalifeh a, Daniel T. Blumstein b,**, Rafaela S. Fontenele a, Kara Schmidlin a, C´ecile Richet a, Simona Kraberger a, Arvind Varsani a,c,* a The Biodesign Center for Fundamental and Applied Microbiomics, School of Life Sciences, Center for Evolution and Medicine, Arizona State University, Tempe, AZ, 85287, USA b Department of Ecology & Evolutionary Biology, Institute of the Environment & Sustainability, University of California Los Angeles, Los Angeles, CA, 90095, USA c Structural Biology Research Unit, Department of Clinical Laboratory Sciences, University of Cape Town, 7925, Cape Town, South Africa ARTICLE INFO ABSTRACT Keywords: Over that last decade, coupling multiple strand displacement approaches with high throughput sequencing have Marmota flaviventer resulted in the identification of genomes of diverse groups of small circular DNA viruses. Using a similar Anelloviridae approach but with recovery of complete genomes by PCR, we identified a diverse group of single-stranded vi Genomoviridae ruses in yellow-bellied marmot (Marmota flaviventer) fecal samples. From 13 fecal samples we identified viruses Cressdnaviricota in the family Genomoviridae (n = 7) and Anelloviridae (n = 1), and several others that ware part of the larger Cressdnaviricota phylum but not within established families (n = 19). There were also circular DNA molecules identified (n = 4) that appear to encode one viral-like gene and have genomes of <1545 nts. This study gives a snapshot of viruses associated with marmots based on fecal sampling. -

Multiple Origins of Prokaryotic and Eukaryotic Single-Stranded DNA Viruses from Bacterial and Archaeal Plasmids
ARTICLE https://doi.org/10.1038/s41467-019-11433-0 OPEN Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids Darius Kazlauskas 1, Arvind Varsani 2,3, Eugene V. Koonin 4 & Mart Krupovic 5 Single-stranded (ss) DNA viruses are a major component of the earth virome. In particular, the circular, Rep-encoding ssDNA (CRESS-DNA) viruses show high diversity and abundance 1234567890():,; in various habitats. By combining sequence similarity network and phylogenetic analyses of the replication proteins (Rep) belonging to the HUH endonuclease superfamily, we show that the replication machinery of the CRESS-DNA viruses evolved, on three independent occa- sions, from the Reps of bacterial rolling circle-replicating plasmids. The CRESS-DNA viruses emerged via recombination between such plasmids and cDNA copies of capsid genes of eukaryotic positive-sense RNA viruses. Similarly, the rep genes of prokaryotic DNA viruses appear to have evolved from HUH endonuclease genes of various bacterial and archaeal plasmids. Our findings also suggest that eukaryotic polyomaviruses and papillomaviruses with dsDNA genomes have evolved via parvoviruses from CRESS-DNA viruses. Collectively, our results shed light on the complex evolutionary history of a major class of viruses revealing its polyphyletic origins. 1 Institute of Biotechnology, Life Sciences Center, Vilnius University, Saulėtekio av. 7, Vilnius 10257, Lithuania. 2 The Biodesign Center for Fundamental and Applied Microbiomics, School of Life Sciences, Center for Evolution and Medicine, Arizona State University, Tempe, AZ 85287, USA. 3 Structural Biology Research Unit, Department of Integrative Biomedical Sciences, University of Cape Town, Rondebosch, 7700 Cape Town, South Africa. -

Primordial Capsid and Spooled Ssdna Genome Structures Penetrate
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.14.435335; this version posted March 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Primordial capsid and spooled ssDNA genome structures penetrate 2 ancestral events of eukaryotic viruses 3 4 Anna Munke1*#, Kei Kimura2, Yuji Tomaru3, Han Wang1, Kazuhiro Yoshida4, Seiya Mito5, Yuki 5 Hongo6, and Kenta Okamoto1* 6 1. The Laboratory of Molecular Biophysics, Department of Cell and Molecular Biology, Uppsala 7 University, Uppsala, Sweden 8 2. Department of Biological Resource Science, Faculty of Agriculture, Saga University, Saga, Japan 9 3. Fisheries Technology Institute, Japan Fisheries Research and Education Agency, Hatsukaichi, 10 Hiroshima, Japan 11 4. Graduate School of Agriculture, Saga University, Saga, Japan 12 5. Department of Biological Resource Science, Faculty of Agriculture, Saga University, Saga, Japan 13 6. Bioinformatics and Biosciences Division, Fisheries Resources Institute, Japan Fisheries Research 14 and Education Agency, Fukuura, Kanazawa, Yokohama, Kanagawa, Japan 15 16 17 *Corresponding authors 18 Corresponding author 1: [email protected] 19 Corresponding author 2: [email protected] 20 21 22 #Present address: Center for Free Electron Laser Science, Deutsches Elektronen-Synchrotron DESY, 23 Hamburg 22607, Germany 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.14.435335; this version posted March 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 24 Abstract 25 Marine algae viruses are important for controlling microorganism communities in the marine 26 ecosystem, and played a fundamental role during the early events of viral evolution. -

Small Circular DNA Viruses: the Muddy Viral “Playground” of Recombinant, Reassortant, and Highly Diverse Viruses
Small Circular DNA Viruses: The muddy viral “playground” of recombinant, reassortant, and highly diverse viruses @Varsani_lab Arizona State University, USA University of Canterbury, New Zealand University of Cape Town, South Africa Arvind Varsani Nucleotide substitutions Recombination Reassortment Viral evolution Viral Nucleotide substitutions Recombination Reassortment Viral evolution Viral Nucleotide substitutions Recombination Reassortment Viral evolution Viral I II III IV V dsDNA ssDNA VI VII dsRNA ssRNA(+) ssRNA(-) ssRNA(RT) dsDNA(RT) viruses Archaea Bacteria Diatoms Fungi Invertebrates Plants Vertebrates Unknown Anelloviridae – circular (~2.1 - 3.8kb) Bacillidnaviridae - circular (~4.5 - 6kb) Circoviridae – circular (~1.6 - 2.3kb) Geminiviridae – circular (~2.7kb; monopartite, bipartite, associated with α/β satellites) Genomovirdae – circular (1.8 -2.3 kb) ssDNA Inoviridae – circular (~5.8 - 8.7kb) Microviridae – circular (~4.5 - 6kb) Nanoviridae – circular (~1.2kb; multicomponent viruses; associated with satellites) Parvoviridae – linear (~3.7 - 6kb) Pleolipoviridae* – circular ssDNA; circular dsDNA or liner dsDNA (7-16kb) Smacoviridae* - circular (2.3 - 2.9kb) ssDNA viruses Chimerism in Rep Geminiviruses MSV: ~2 x 10-4 subs/site/year SSRV: ~3 x 10-4 subs/site/year EACMV: ~1 x 10-3 subs/site/year TYLCV: ~3 x 10-4 subs/site/year Nanoviruses FBNYV: ~1 x 10-3 subs/site/year BBTV: ~3 x 10-4 subs/site/year Circoviruses BFDV: ~2 x 10-3 subs/site/year PCV-2: ~1 x 10-3 subs/site/year Parvoviruses CPV: ~2 x 10-4 - 8 x 10-5 subs/site/year -

Beet Curly Top Virus Strains Associated with Sugar Beet in Idaho, Oregon, and a Western U.S
Plant Disease • 2017 • 101:1373-1382 • http://dx.doi.org/10.1094/PDIS-03-17-0381-RE Beet curly top virus Strains Associated with Sugar Beet in Idaho, Oregon, and a Western U.S. Collection Carl A. Strausbaugh and Imad A. Eujayl, United States Department of Agriculture–Agricultural Research Service (USDA-ARS) Northwest Irrigation and Soils Research Laboratory, Kimberly, ID 83341; and William M. Wintermantel, USDA-ARS, Salinas, CA 93905 Abstract Curly top of sugar beet is a serious, yield-limiting disease in semiarid pro- Logan) strains and primers that amplified a group of Worland (Wor)- duction areas caused by Beet curly top virus (BCTV) and transmitted like strains. The BCTV strain distribution averaged 2% Svr, 30% CA/ by the beet leafhopper. One of the primary means of control for BCTV Logan, and 87% Wor-like (16% had mixed infections), which differed in sugar beet is host resistance but effectiveness of resistance can vary from the previously published 2006-to-2007 collection (87% Svr, 7% among BCTV strains. Strain prevalence among BCTV populations CA/Logan, and 60% Wor-like; 59% mixed infections) based on a contin- was last investigated in Idaho and Oregon during a 2006-to-2007 collec- gency test (P < 0.0001). Whole-genome sequencing (GenBank acces- tion but changes in disease severity suggested a need for reevaluation. sions KT276895 to KT276920 and KX867015 to KX867057) with Therefore, 406 leaf samples symptomatic for curly top were collected overlapping primers found that the Wor-like strains included Wor, Colo- from sugar beet plants in commercial sugar beet fields in Idaho and rado and a previously undescribed strain designated Kimberly1. -

Geminiviruses Encode Additional Small Proteins with Specific
ARTICLE https://doi.org/10.1038/s41467-021-24617-4 OPEN Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function Pan Gong 1,6, Huang Tan 2,3,6, Siwen Zhao1, Hao Li1, Hui Liu4,YuMa2,3, Xi Zhang2,3, Junjie Rong2,3, Xing Fu2, ✉ ✉ ✉ Rosa Lozano-Durán 2,5 , Fangfang Li1 & Xueping Zhou 1,4 1234567890():,; Geminiviruses are plant viruses with limited coding capacity. Geminivirus-encoded proteins are traditionally identified by applying a 10-kDa arbitrary threshold; however, it is increasingly clear that small proteins play relevant roles in biological systems, which calls for the reconsideration of this criterion. Here, we show that geminiviral genomes contain additional ORFs. Using tomato yellow leaf curl virus, we demonstrate that some of these small ORFs are expressed during the infection, and that the encoded proteins display specific subcellular localizations. We prove that the largest of these additional ORFs, which we name V3, is required for full viral infection, and that the V3 protein localizes in the Golgi apparatus and functions as an RNA silencing suppressor. These results imply that the repertoire of gemi- niviral proteins can be expanded, and that getting a comprehensive overview of the molecular plant-geminivirus interactions will require the detailed study of small ORFs so far neglected. 1 State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China. 2 Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China. 3 University of the Chinese Academy of Sciences, Beijing, China. -

Viral Diversity in Tree Species
Universidade de Brasília Instituto de Ciências Biológicas Departamento de Fitopatologia Programa de Pós-Graduação em Biologia Microbiana Doctoral Thesis Viral diversity in tree species FLÁVIA MILENE BARROS NERY Brasília - DF, 2020 FLÁVIA MILENE BARROS NERY Viral diversity in tree species Thesis presented to the University of Brasília as a partial requirement for obtaining the title of Doctor in Microbiology by the Post - Graduate Program in Microbiology. Advisor Dra. Rita de Cássia Pereira Carvalho Co-advisor Dr. Fernando Lucas Melo BRASÍLIA, DF - BRAZIL FICHA CATALOGRÁFICA NERY, F.M.B Viral diversity in tree species Flávia Milene Barros Nery Brasília, 2025 Pages number: 126 Doctoral Thesis - Programa de Pós-Graduação em Biologia Microbiana, Universidade de Brasília, DF. I - Virus, tree species, metagenomics, High-throughput sequencing II - Universidade de Brasília, PPBM/ IB III - Viral diversity in tree species A minha mãe Ruth Ao meu noivo Neil Dedico Agradecimentos A Deus, gratidão por tudo e por ter me dado uma família e amigos que me amam e me apoiam em todas as minhas escolhas. Minha mãe Ruth e meu noivo Neil por todo o apoio e cuidado durante os momentos mais difíceis que enfrentei durante minha jornada. Aos meus irmãos André, Diego e meu sobrinho Bruno Kawai, gratidão. Aos meus amigos de longa data Rafaelle, Evanessa, Chênia, Tati, Leo, Suzi, Camilets, Ricardito, Jorgito e Diego, saudade da nossa amizade e dos bons tempos. Amo vocês com todo o meu coração! Minha orientadora e grande amiga Profa Rita de Cássia Pereira Carvalho, a quem escolhi e fui escolhida para amar e fazer parte da família. -
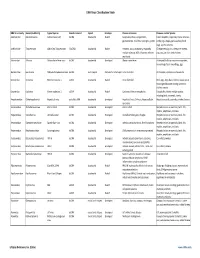
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Viruses in Transplantation - Not Always Enemies
Viruses in transplantation - not always enemies Virome and transplantation ECCMID 2018 - Madrid Prof. Laurent Kaiser Head Division of Infectious Diseases Laboratory of Virology Geneva Center for Emerging Viral Diseases University Hospital of Geneva ESCMID eLibrary © by author Conflict of interest None ESCMID eLibrary © by author The human virome: definition? Repertoire of viruses found on the surface of/inside any body fluid/tissue • Eukaryotic DNA and RNA viruses • Prokaryotic DNA and RNA viruses (phages) 25 • The “main” viral community (up to 10 bacteriophages in humans) Haynes M. 2011, Metagenomic of the human body • Endogenous viral elements integrated into host chromosomes (8% of the human genome) • NGS is shaping the definition Rascovan N et al. Annu Rev Microbiol 2016;70:125-41 Popgeorgiev N et al. Intervirology 2013;56:395-412 Norman JM et al. Cell 2015;160:447-60 ESCMID eLibraryFoxman EF et al. Nat Rev Microbiol 2011;9:254-64 © by author Viruses routinely known to cause diseases (non exhaustive) Upper resp./oropharyngeal HSV 1 Influenza CNS Mumps virus Rhinovirus JC virus RSV Eye Herpes viruses Parainfluenza HSV Measles Coronavirus Adenovirus LCM virus Cytomegalovirus Flaviviruses Rabies HHV6 Poliovirus Heart Lower respiratory HTLV-1 Coxsackie B virus Rhinoviruses Parainfluenza virus HIV Coronaviruses Respiratory syncytial virus Parainfluenza virus Adenovirus Respiratory syncytial virus Coronaviruses Gastro-intestinal Influenza virus type A and B Human Bocavirus 1 Adenovirus Hepatitis virus type A, B, C, D, E Those that cause -

Ocorrência Natural De Espécies Virais E Avaliação De Potenciais Hospedeiras Experimentais De Begomovírus, Potyvírus E Tospovírus Em Plantas Arbóreas E Berinjela
UNIVERSIDADE DE BRASÍLIA INSTITUTO DE CIÊNCIAS BIOLÓGICAS DEPARTAMENTO DE FITOPATOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM FITOPATOLOGIA Ocorrência natural de espécies virais e avaliação de potenciais hospedeiras experimentais de begomovírus, potyvírus e tospovírus em plantas arbóreas e berinjela (Solanum melongena) JOSIANE GOULART BATISTA Brasília - DF 2014 JOSIANE GOULART BATISTA Ocorrência natural de espécies virais e avaliação de potenciais hospedeiras experimentais de begomovírus, potyvírus e tospovírus em plantas arbóreas e berinjela (Solanum melongena) Dissertação apresentada à Universidade de Brasília como requisito parcial para a obtenção do título de Mestre em Fitopatologia pelo Programa de Pós Graduação em Fitopatologia. Orientadora Dra. Rita de Cássia Pereira Carvalho BRASÍLIA DISTRITO FEDERAL – BRASIL 2014 FICHA CATALOGRÁFICA Batista, G.J. Ocorrência natural de espécies virais e avaliação de potenciais hospedeiras experimentais de begomovírus, potyvírus e tospovírus em plantas arbóreas e berinjela (Solanum melongena). Josiane Goulart Batista. Brasília, 2014 Número de páginas p.: 183 Dissertação de mestrado - Programa de Pós-Graduação em Fitopatologia, Universidade de Brasília, Brasília. Árvores, berinjela, vírus, resistência. I. Universidade de Brasília. PPG/FIT. II. Título. Ocorrência natural de espécies virais e avaliação de potenciais hospedeiras experimentais de begomovírus, potyvírus e tospovírus em plantas arbóreas e berinjela (Solanum melongena). Dedico a minha pequena linda filha Giovanna, ao meu pai Josvaldo, a minha mãe Nelma, ao meu namorado Airton, a minha irmã Josefa e minha sobrinha Júlia. Agradecimentos A Deus. Aos meus pais Josvaldo e Nelma. A minha filha Giovanna, minha irmã Josefa e minha sobrinha Júlia. A minha orientadora Professora Rita de Cássia. Aos colegas do mestrado e doutorado Rafaela, Marcella, Juliana, Ícaro, Cléia, Monica, Elenice, William, Rayane e Nédio. -

Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics
viruses Article Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics Ashleigh F. Porter 1, Mang Shi 1, John-Sebastian Eden 1,2 , Yong-Zhen Zhang 3,4 and Edward C. Holmes 1,3,* 1 Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life & Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia; [email protected] (A.F.P.); [email protected] (M.S.); [email protected] (J.-S.E.) 2 Centre for Virus Research, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia 3 Shanghai Public Health Clinical Center and School of Public Health, Fudan University, Shanghai 201500, China; [email protected] 4 Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China * Correspondence: [email protected]; Tel.: +61-2-9351-5591 Received: 17 October 2019; Accepted: 23 November 2019; Published: 25 November 2019 Abstract: DNA viruses comprise a wide array of genome structures and infect diverse host species. To date, most studies of DNA viruses have focused on those with the strongest disease associations. Accordingly, there has been a marked lack of sampling of DNA viruses from invertebrates. Bulk RNA sequencing has resulted in the discovery of a myriad of novel RNA viruses, and herein we used this methodology to identify actively transcribing DNA viruses in meta-transcriptomic libraries of diverse invertebrate species. Our analysis revealed high levels of phylogenetic diversity in DNA viruses, including 13 species from the Parvoviridae, Circoviridae, and Genomoviridae families of single-stranded DNA virus families, and six double-stranded DNA virus species from the Nudiviridae, Polyomaviridae, and Herpesviridae, for which few invertebrate viruses have been identified to date.