Phylogenetics of Liliales
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE
Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE LILIACEAE de Jussieu 1789 (Lily Family) (also see AGAVACEAE, ALLIACEAE, ALSTROEMERIACEAE, AMARYLLIDACEAE, ASPARAGACEAE, COLCHICACEAE, HEMEROCALLIDACEAE, HOSTACEAE, HYACINTHACEAE, HYPOXIDACEAE, MELANTHIACEAE, NARTHECIACEAE, RUSCACEAE, SMILACACEAE, THEMIDACEAE, TOFIELDIACEAE) As here interpreted narrowly, the Liliaceae constitutes about 11 genera and 550 species, of the Northern Hemisphere. There has been much recent investigation and re-interpretation of evidence regarding the upper-level taxonomy of the Liliales, with strong suggestions that the broad Liliaceae recognized by Cronquist (1981) is artificial and polyphyletic. Cronquist (1993) himself concurs, at least to a degree: "we still await a comprehensive reorganization of the lilies into several families more comparable to other recognized families of angiosperms." Dahlgren & Clifford (1982) and Dahlgren, Clifford, & Yeo (1985) synthesized an early phase in the modern revolution of monocot taxonomy. Since then, additional research, especially molecular (Duvall et al. 1993, Chase et al. 1993, Bogler & Simpson 1995, and many others), has strongly validated the general lines (and many details) of Dahlgren's arrangement. The most recent synthesis (Kubitzki 1998a) is followed as the basis for familial and generic taxonomy of the lilies and their relatives (see summary below). References: Angiosperm Phylogeny Group (1998, 2003); Tamura in Kubitzki (1998a). Our “liliaceous” genera (members of orders placed in the Lilianae) are therefore divided as shown below, largely following Kubitzki (1998a) and some more recent molecular analyses. ALISMATALES TOFIELDIACEAE: Pleea, Tofieldia. LILIALES ALSTROEMERIACEAE: Alstroemeria COLCHICACEAE: Colchicum, Uvularia. LILIACEAE: Clintonia, Erythronium, Lilium, Medeola, Prosartes, Streptopus, Tricyrtis, Tulipa. MELANTHIACEAE: Amianthium, Anticlea, Chamaelirium, Helonias, Melanthium, Schoenocaulon, Stenanthium, Veratrum, Toxicoscordion, Trillium, Xerophyllum, Zigadenus. -

Alphabetical Lists of the Vascular Plant Families with Their Phylogenetic
Colligo 2 (1) : 3-10 BOTANIQUE Alphabetical lists of the vascular plant families with their phylogenetic classification numbers Listes alphabétiques des familles de plantes vasculaires avec leurs numéros de classement phylogénétique FRÉDÉRIC DANET* *Mairie de Lyon, Espaces verts, Jardin botanique, Herbier, 69205 Lyon cedex 01, France - [email protected] Citation : Danet F., 2019. Alphabetical lists of the vascular plant families with their phylogenetic classification numbers. Colligo, 2(1) : 3- 10. https://perma.cc/2WFD-A2A7 KEY-WORDS Angiosperms family arrangement Summary: This paper provides, for herbarium cura- Gymnosperms Classification tors, the alphabetical lists of the recognized families Pteridophytes APG system in pteridophytes, gymnosperms and angiosperms Ferns PPG system with their phylogenetic classification numbers. Lycophytes phylogeny Herbarium MOTS-CLÉS Angiospermes rangement des familles Résumé : Cet article produit, pour les conservateurs Gymnospermes Classification d’herbier, les listes alphabétiques des familles recon- Ptéridophytes système APG nues pour les ptéridophytes, les gymnospermes et Fougères système PPG les angiospermes avec leurs numéros de classement Lycophytes phylogénie phylogénétique. Herbier Introduction These alphabetical lists have been established for the systems of A.-L de Jussieu, A.-P. de Can- The organization of herbarium collections con- dolle, Bentham & Hooker, etc. that are still used sists in arranging the specimens logically to in the management of historical herbaria find and reclassify them easily in the appro- whose original classification is voluntarily pre- priate storage units. In the vascular plant col- served. lections, commonly used methods are systema- Recent classification systems based on molecu- tic classification, alphabetical classification, or lar phylogenies have developed, and herbaria combinations of both. -

Draft Plant Propagation Protocol
Plant Propagation Protocol for Zigadenus elegans ESRM 412 – Native Plant Production TAXONOMY Family Names Family Scientific Name: Liliaceae Family Common Name: Lily family Scientific Names Genus: Zigadenus Michx. Species: Zigadenus elegans Species Authority: Pursh Variety: Sub-species: Zigadenus elegans ssp. elegans Zigadenus elegans ssp. glaucus Cultivar: Authority for Variety/Sub-species: Common Synonym(s) Anticlea coloradensis (Rydb.) Rydb. Anticlea elegans (Pursh) Rydb. Zigadenus alpinus Blank. Zigadenus elegans Pursh ssp. elegans 2 Common Name(s): Glaucous death camas, Mountain death camas, White camas 2 Species Code : ZIEL2 GENERAL INFORMATION Geographical range See above 1 Ecological distribution : Occurs in meadows, open forests and rocky slopes, at middle to high elevations in the mountains 2 Other sources indicate it can also be found in moist grasslands, river and lake shores, and bogs in coniferous forests. 6 9 It has also been listed as an indicator species for areas that have been former savanna's/woodlands. Climate and elevation range Subalpine meadows and moist screes at high elevations in the Rockies and Pacific Coast states. 12 Local habitat and abundance; may Occurs in sandy, moist soils. It can tolerate partial include commonly associated shade but also needs sunlight. 5 species It and other indicator species tend to be strongly limited to partial canopy conditions. In more heavily-wooded sites, these species are usually in a state of decline due to the increasing canopy closure above. They are therefore dependent on canopy gaps, edges, roadsides etc. in densely-wooded areas. 9 In Missouri it cam be found on the crevices and ledges of north-facing dolomite bluffs. -

Tracing History
Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 911 Tracing History Phylogenetic, Taxonomic, and Biogeographic Research in the Colchicum Family BY ANNIKA VINNERSTEN ACTA UNIVERSITATIS UPSALIENSIS UPPSALA 2003 Dissertation presented at Uppsala University to be publicly examined in Lindahlsalen, EBC, Uppsala, Friday, December 12, 2003 at 10:00 for the degree of Doctor of Philosophy. The examination will be conducted in English. Abstract Vinnersten, A. 2003. Tracing History. Phylogenetic, Taxonomic and Biogeographic Research in the Colchicum Family. Acta Universitatis Upsaliensis. Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 911. 33 pp. Uppsala. ISBN 91-554-5814-9 This thesis concerns the history and the intrafamilial delimitations of the plant family Colchicaceae. A phylogeny of 73 taxa representing all genera of Colchicaceae, except the monotypic Kuntheria, is presented. The molecular analysis based on three plastid regions—the rps16 intron, the atpB- rbcL intergenic spacer, and the trnL-F region—reveal the intrafamilial classification to be in need of revision. The two tribes Iphigenieae and Uvularieae are demonstrated to be paraphyletic. The well-known genus Colchicum is shown to be nested within Androcymbium, Onixotis constitutes a grade between Neodregea and Wurmbea, and Gloriosa is intermixed with species of Littonia. Two new tribes are described, Burchardieae and Tripladenieae, and the two tribes Colchiceae and Uvularieae are emended, leaving four tribes in the family. At generic level new combinations are made in Wurmbea and Gloriosa in order to render them monophyletic. The genus Androcymbium is paraphyletic in relation to Colchicum and the latter genus is therefore expanded. -

Complete Chloroplast Genomes Shed Light on Phylogenetic
www.nature.com/scientificreports OPEN Complete chloroplast genomes shed light on phylogenetic relationships, divergence time, and biogeography of Allioideae (Amaryllidaceae) Ju Namgung1,4, Hoang Dang Khoa Do1,2,4, Changkyun Kim1, Hyeok Jae Choi3 & Joo‑Hwan Kim1* Allioideae includes economically important bulb crops such as garlic, onion, leeks, and some ornamental plants in Amaryllidaceae. Here, we reported the complete chloroplast genome (cpDNA) sequences of 17 species of Allioideae, fve of Amaryllidoideae, and one of Agapanthoideae. These cpDNA sequences represent 80 protein‑coding, 30 tRNA, and four rRNA genes, and range from 151,808 to 159,998 bp in length. Loss and pseudogenization of multiple genes (i.e., rps2, infA, and rpl22) appear to have occurred multiple times during the evolution of Alloideae. Additionally, eight mutation hotspots, including rps15-ycf1, rps16-trnQ-UUG, petG-trnW-CCA , psbA upstream, rpl32- trnL-UAG , ycf1, rpl22, matK, and ndhF, were identifed in the studied Allium species. Additionally, we present the frst phylogenomic analysis among the four tribes of Allioideae based on 74 cpDNA coding regions of 21 species of Allioideae, fve species of Amaryllidoideae, one species of Agapanthoideae, and fve species representing selected members of Asparagales. Our molecular phylogenomic results strongly support the monophyly of Allioideae, which is sister to Amaryllioideae. Within Allioideae, Tulbaghieae was sister to Gilliesieae‑Leucocoryneae whereas Allieae was sister to the clade of Tulbaghieae‑ Gilliesieae‑Leucocoryneae. Molecular dating analyses revealed the crown age of Allioideae in the Eocene (40.1 mya) followed by diferentiation of Allieae in the early Miocene (21.3 mya). The split of Gilliesieae from Leucocoryneae was estimated at 16.5 mya. -
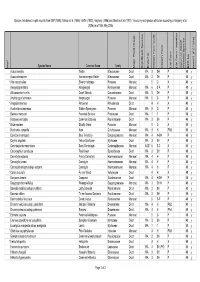
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Nordic Journal of Botany NJB-02477 She, R., Zhao, P
Nordic Journal of Botany NJB-02477 She, R., Zhao, P. , Zhou, H., Yue, M., Yan, F., Hu, G., Gao, X. and Zhang, S. 2020. Complete chloroplast genomes of Liliaceae (s.l.) species: comparative genomic and phylogenetic analyses. - Nordic Journal of Botany 2019: e02477 Appendix 1 Table A1. List of taxa sampled in this study and species accessions numbers (GenBank). Genus Species Accession number Genus Species Accession number Aletris fauriei KT898912 Polygonatum sibiricum KT695605 Aletris Polygonatum Aletris spicata KT898911 Polygonatum verticillatum KT722981 Allium cepa KM088013 Lilium amabile KY940845 Allium obliquum MG670111 Lilium bakerianum KY748301 Allium Allium prattii MG739457 Lilium brownii KY748296 Allium victorialis MF687749 Lilium bulbiferum MG574829 Amana anhuiensis KY401423 Lilium callosum KY940846 Amana edulis KY401425 Lilium cernuum KX354692 Amana Amana erythronioides KY401424 Lilium distichum KT376489 Amana kuocangshanica KY401426 Lilium duchartrei KY748300 Amana wanzhensis KY401422 Lilium fargesii KX592156 Fritillaria cirrhosa KF769143 Lilium hansonii KM103364 Lilium Fritillaria eduardii MF947708 Lilium henryi KY748302 Fritillaria hupehensis KF712486 Lilium lancifolium KY748297 Fritillaria meleagroides MF947710 Lilium leucanthum KY748299 Fritillaria persica MF947709 Lilium longiflorum KC968977 Fritillaria Fritillaria thunbergii KY646165 Lilium philadelphicum KY940847 Fritillaria unibracteata var. KF769142 Lilium primulinum var. ochraceum KY748298 wabuensis Fritillaria ussuriensis KY646166 Lilium taliense KY009938 Fritillaria -

Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University
Aliso: A Journal of Systematic and Evolutionary Botany Volume 22 | Issue 1 Article 3 2006 Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University Thomas Janssen Muséum National d'Histoire Naturelle Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Botany Commons Recommended Citation Bremer, Kåre and Janssen, Thomas (2006) "Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 22: Iss. 1, Article 3. Available at: http://scholarship.claremont.edu/aliso/vol22/iss1/3 Aliso 22, pp. 22-27 © 2006, Rancho Santa Ana Botanic Garden GONDWANAN ORIGIN OF MAJOR MONO COT GROUPS INFERRED FROM DISPERSAL-VICARIANCE ANALYSIS KARE BREMERl.3 AND THOMAS JANSSEN2 lDepartment of Systematic Botany, Evolutionary Biology Centre, Norbyvagen l8D, SE-752 36 Uppsala, Sweden; 2Museum National d'Histoire Naturelle, Departement de Systematique et Evolution, USM 0602: Taxonomie et collections, 16 rue Buffon, 75005 Paris, France 3Corresponding author ([email protected]) ABSTRACT Historical biogeography of major monocot groups was investigated by biogeographical analysis of a dated phylogeny including 79 of the 81 monocot families using the Angiosperm Phylogeny Group II (APG II) classification. Five major areas were used to describe the family distributions: Eurasia, North America, South America, Africa including Madagascar, and Australasia including New Guinea, New Caledonia, and New Zealand. In order to investigate the possible correspondence with continental breakup, the tree with its terminal distributions was fitted to the geological area cladogram «Eurasia, North America), (Africa, (South America, Australasia») and to alternative area cladograms using the TreeFitter program. -

Biodiversity and Ecology of Critically Endangered, Rûens Silcrete Renosterveld in the Buffeljagsrivier Area, Swellendam
Biodiversity and Ecology of Critically Endangered, Rûens Silcrete Renosterveld in the Buffeljagsrivier area, Swellendam by Johannes Philippus Groenewald Thesis presented in fulfilment of the requirements for the degree of Masters in Science in Conservation Ecology in the Faculty of AgriSciences at Stellenbosch University Supervisor: Prof. Michael J. Samways Co-supervisor: Dr. Ruan Veldtman December 2014 Stellenbosch University http://scholar.sun.ac.za Declaration I hereby declare that the work contained in this thesis, for the degree of Master of Science in Conservation Ecology, is my own work that have not been previously published in full or in part at any other University. All work that are not my own, are acknowledge in the thesis. ___________________ Date: ____________ Groenewald J.P. Copyright © 2014 Stellenbosch University All rights reserved ii Stellenbosch University http://scholar.sun.ac.za Acknowledgements Firstly I want to thank my supervisor Prof. M. J. Samways for his guidance and patience through the years and my co-supervisor Dr. R. Veldtman for his help the past few years. This project would not have been possible without the help of Prof. H. Geertsema, who helped me with the identification of the Lepidoptera and other insect caught in the study area. Also want to thank Dr. K. Oberlander for the help with the identification of the Oxalis species found in the study area and Flora Cameron from CREW with the identification of some of the special plants growing in the area. I further express my gratitude to Dr. Odette Curtis from the Overberg Renosterveld Project, who helped with the identification of the rare species found in the study area as well as information about grazing and burning of Renosterveld. -

Redalyc.A Conspectus of Mexican Melanthiaceae Including A
Acta Botánica Mexicana ISSN: 0187-7151 [email protected] Instituto de Ecología, A.C. México Frame, Dawn; Espejo, Adolfo; López Ferrari, Ana Rosa A conspectus of mexican Melanthiaceae including a description of new taxa of schoenocaulon and Zigadenus Acta Botánica Mexicana, núm. 48, septiembre, 1999, pp. 27 - 50 Instituto de Ecología, A.C. Pátzcuaro, México Disponible en: http://www.redalyc.org/articulo.oa?id=57404804 Cómo citar el artículo Número completo Sistema de Información Científica Más información del artículo Red de Revistas Científicas de América Latina, el Caribe, España y Portugal Página de la revista en redalyc.org Proyecto académico sin fines de lucro, desarrollado bajo la iniciativa de acceso abierto Acta Botánica Mexicana (1999), 48:27-50 A CONSPECTUS OF MEXICAN MELANTHIACEAE INCLUDING A DESCRIPTION OF NEW TAXA OF SCHOENOCAULON AND ZIGADENUS DAWN FRAME Laboratoire de Botanique, ISEM Institut de Botanique 163, rue A. Broussonet 34090 Montpellier France e-mail: [email protected] ADOLFO ESPEJO Y ANA ROSA LOPEZ-FERRARI Herbario Metropolitano Departamento de Biología, CBS Universidad Autónoma Metropolitana Unidad Iztapalapa Apartado postal 55-535 09340 México, D.F. e-mail: [email protected] ABSTRACT Seven new taxa of Schoenocaulon and one more of Zigadenus from Mexico are herein described. In addition, a brief description of the Mexican genera of Melanthiaceae and a key to genera and species known from Mexico are given. RESUMEN Se describen siete nuevos taxa de Schoenocaulon y uno de Zigadenus provenientes de diversos estados de México. Se incluyen claves para la identificación de los géneros y de las especies de la familia presentes en México y se hacen algunos comentarios breves sobre los mismos. -

GENOME EVOLUTION in MONOCOTS a Dissertation
GENOME EVOLUTION IN MONOCOTS A Dissertation Presented to The Faculty of the Graduate School At the University of Missouri In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy By Kate L. Hertweck Dr. J. Chris Pires, Dissertation Advisor JULY 2011 The undersigned, appointed by the dean of the Graduate School, have examined the dissertation entitled GENOME EVOLUTION IN MONOCOTS Presented by Kate L. Hertweck A candidate for the degree of Doctor of Philosophy And hereby certify that, in their opinion, it is worthy of acceptance. Dr. J. Chris Pires Dr. Lori Eggert Dr. Candace Galen Dr. Rose‐Marie Muzika ACKNOWLEDGEMENTS I am indebted to many people for their assistance during the course of my graduate education. I would not have derived such a keen understanding of the learning process without the tutelage of Dr. Sandi Abell. Members of the Pires lab provided prolific support in improving lab techniques, computational analysis, greenhouse maintenance, and writing support. Team Monocot, including Dr. Mike Kinney, Dr. Roxi Steele, and Erica Wheeler were particularly helpful, but other lab members working on Brassicaceae (Dr. Zhiyong Xiong, Dr. Maqsood Rehman, Pat Edger, Tatiana Arias, Dustin Mayfield) all provided vital support as well. I am also grateful for the support of a high school student, Cady Anderson, and an undergraduate, Tori Docktor, for their assistance in laboratory procedures. Many people, scientist and otherwise, helped with field collections: Dr. Travis Columbus, Hester Bell, Doug and Judy McGoon, Julie Ketner, Katy Klymus, and William Alexander. Many thanks to Barb Sonderman for taking care of my greenhouse collection of many odd plants brought back from the field. -

Pollen Morphology of Erythronium L. (Liliaceae) and Its Systematic Relationships
J. Basic. Appl. Sci. Res., 2(2)1833-1838, 2012 ISSN 2090-4304 Journal of Basic and Applied © 2012, TextRoad Publication Scientific Research www.textroad.com Pollen Morphology of Erythronium L. (Liliaceae) and its Systematic Relationships Sayed-Mohammad Masoumi Department of Plant Protection, Razi University, Kermanshah, Iran ABSTRACT Pollen morphology of three genus of Erythronium was studied by the Light Microscopy (LM), Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM). Sulcus long reaching the ends of the grains, with operculum (E. giganteum, E. sibiricum) or without it (E. caucasicum). With surface latticed ornamentation and large lattice, thickness of muri and size of Lumina in E. sibiricum are widely varied. Also, most palynomorphological characteristics of the data transmission electron microscopy (TEM) showed no strong differences between E. caucasicum and E. sibiricum, , but these species are well distinguished from E. giganteum according to ectexine thickness (thickness of the tectum and the foot layer), shape and diameter of the caput, height and width of the columella. KEY WORDS: Caput; Columella; Exine ornamentation; intine; Microrelief; Pollen grain; Tectum. INTRODUCTION Takhtajan, 1987 indicated that the genus of Erythronium in Tribe Tulipeae is of the Liliaceae family. Different sources have considered the species number of this genus varied from 24-30. Baranova (1999) introduced 24 species for this genus, of which 20 species were spread in North America. Allen et al. (2003) examined the genus of Erythronium, Amana, and Tulipa using the DNA sequences from the chloroplast gene matK and the internal transcribed spacer (ITS) of nuclear ribosomal DNA. Palynomorphological characters of 20 different pollen species of Erythronium were evaluated by different researchers (Ikuse, 1965; Beug (1963); Radulescu, 1973; Nakamura, 1980; Schulze, 1980; Kuprianova, 1983; Takahashi (1987); Kosenko, 1991b, 1992, 1996, 1999; Maassoumi, 2005a, 2005b, 2007).