MEMORIAL SLOAN-KETTERING CANCER CENTER Curriculum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
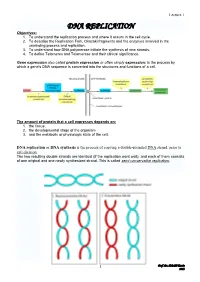
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

Indications for a Central Role of Hexokinase Activity in Natural Variation of Heat Acclimation in Arabidopsis Thaliana
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 June 2020 doi:10.20944/preprints202006.0169.v1 Article Indications for a central role of hexokinase activity in natural variation of heat acclimation in Arabidopsis thaliana Vasil Atanasov §, Lisa Fürtauer § and Thomas Nägele * LMU Munich, Plant Evolutionary Cell Biology, Großhaderner Str. 2-4, 82152 Planegg, Germany § Authors contributed equally * Correspondence: [email protected] Abstract: Diurnal and seasonal changes of abiotic environmental factors shape plant performance and distribution. Changes of growth temperature and light intensity may vary significantly on a diurnal, but also on a weekly or seasonal scale. Hence, acclimation to a changing temperature and light regime is essential for plant survival and propagation. In the present study, we analyzed photosynthetic CO2 assimilation and metabolic regulation of the central carbohydrate metabolism in two natural accessions of Arabidopsis thaliana originating from Russia and south Italy during exposure to heat and a combination of heat and high light. Our findings indicate that it is hardly possible to predict photosynthetic capacities to fix CO2 under combined stress from single stress experiments. Further, capacities of hexose phosphorylation were found to be significantly lower in the Italian than in the Russian accession which could explain an inverted sucrose-to-hexose ratio. Together with the finding of significantly stronger accumulation of anthocyanins under heat/high light these observations indicate a central role of hexokinase activity in stabilization of photosynthetic capacities within a changing environment. Keywords: photosynthesis; carbohydrate metabolism; hexokinase; heat acclimation; environmental changes; natural variation; high light; combined stress. 1. Introduction Changes of growth temperature and light intensity broadly affect plant molecular, physiological and developmental processes. -

METABOLIC EVOLUTION in GALDIERIA SULPHURARIA By
METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA By CHAD M. TERNES Bachelor of Science in Botany Oklahoma State University Stillwater, Oklahoma 2009 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY May, 2015 METABOLIC EVOLUTION IN GALDIERIA SUPHURARIA Dissertation Approved: Dr. Gerald Schoenknecht Dissertation Adviser Dr. David Meinke Dr. Andrew Doust Dr. Patricia Canaan ii Name: CHAD M. TERNES Date of Degree: MAY, 2015 Title of Study: METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA Major Field: PLANT SCIENCE Abstract: The thermoacidophilic, unicellular, red alga Galdieria sulphuraria possesses characteristics, including salt and heavy metal tolerance, unsurpassed by any other alga. Like most plastid bearing eukaryotes, G. sulphuraria can grow photoautotrophically. Additionally, it can also grow solely as a heterotroph, which results in the cessation of photosynthetic pigment biosynthesis. The ability to grow heterotrophically is likely correlated with G. sulphuraria ’s broad capacity for carbon metabolism, which rivals that of fungi. Annotation of the metabolic pathways encoded by the genome of G. sulphuraria revealed several pathways that are uncharacteristic for plants and algae, even red algae. Phylogenetic analyses of the enzymes underlying the metabolic pathways suggest multiple instances of horizontal gene transfer, in addition to endosymbiotic gene transfer and conservation through ancestry. Although some metabolic pathways as a whole appear to be retained through ancestry, genes encoding individual enzymes within a pathway were substituted by genes that were acquired horizontally from other domains of life. Thus, metabolic pathways in G. sulphuraria appear to be composed of a ‘metabolic patchwork’, underscored by a mosaic of genes resulting from multiple evolutionary processes. -

Biochemistrystanford00kornrich.Pdf
University of California Berkeley Regional Oral History Office University of California The Bancroft Library Berkeley, California Program in the History of the Biosciences and Biotechnology Arthur Kornberg, M.D. BIOCHEMISTRY AT STANFORD, BIOTECHNOLOGY AT DNAX With an Introduction by Joshua Lederberg Interviews Conducted by Sally Smith Hughes, Ph.D. in 1997 Copyright 1998 by The Regents of the University of California Since 1954 the Regional Oral History Office has been interviewing leading participants in or well-placed witnesses to major events in the development of Northern California, the West, and the Nation. Oral history is a method of collecting historical information through tape-recorded interviews between a narrator with firsthand knowledge of historically significant events and a well- informed interviewer, with the goal of preserving substantive additions to the historical record. The tape recording is transcribed, lightly edited for continuity and clarity, and reviewed by the interviewee. The corrected manuscript is indexed, bound with photographs and illustrative materials, and placed in The Bancroft Library at the University of California, Berkeley, and in other research collections for scholarly use. Because it is primary material, oral history is not intended to present the final, verified, or complete narrative of events. It is a spoken account, offered by the interviewee in response to questioning, and as such it is reflective, partisan, deeply involved, and irreplaceable. ************************************ All uses of this manuscript are covered by a legal agreement between The Regents of the University of California and Arthur Kornberg, M.D., dated June 18, 1997. The manuscript is thereby made available for research purposes. All literary rights in the manuscript, including the right to publish, are reserved to The Bancroft Library of the University of California, Berkeley. -

Liver Med23 Ablation Improves Glucose and Lipid Metabolism Through Modulating FOXO1 Activity
Cell Research (2014) 24:1250-1265. npg © 2014 IBCB, SIBS, CAS All rights reserved 1001-0602/14 ORIGINAL ARTICLE www.nature.com/cr Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity Yajing Chu1, Leonardo Gómez Rosso1, Ping Huang2, Zhichao Wang1, Yichi Xu3, Xiao Yao1, Menghan Bao1, Jun Yan3, Haiyun Song2, Gang Wang1 1State Key Laboratory of Cell Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chi- nese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China; 2Key Laboratory of Nutrition and Metabolism, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shang- hai 200031, China; 3CAS-MPG Partner Institute for Computational Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China Mediator complex is a molecular hub integrating signaling, transcription factors, and RNA polymerase II (RNAPII) machinery. Mediator MED23 is involved in adipogenesis and smooth muscle cell differentiation, suggesting its role in energy homeostasis. Here, through the generation and analysis of a liver-specific Med23-knockout mouse, we found that liver Med23 deletion improved glucose and lipid metabolism, as well as insulin responsiveness, and prevented diet-induced obesity. Remarkably, acute hepatic Med23 knockdown in db/db mice significantly improved the lipid profile and glucose tolerance. Mechanistically, MED23 participates in gluconeogenesis and cholesterol synthesis through modulating the transcriptional activity of FOXO1, a key metabolic transcription factor. Indeed, hepatic Med23 deletion impaired the Mediator and RNAPII recruitment and attenuated the expression of FOXO1 target genes. Moreover, this functional interaction between FOXO1 and MED23 is evolutionarily conserved, as the in vivo activities of dFOXO in larval fat body and in adult wing can be partially blocked by Med23 knockdown in Drosophila. -

2012 Annual Report
Memorial Sloan-Kettering Cancer Center 2012 ANNUAL REPORT A SHARED VISION A SINGULAR MISSION Nurse practitioner Naomi Cazeau, of the Adult Bone Marrow Transplant Service. PING CHI PHYSICIAN-SCIENTIST 10 STEPHEN SOLOMON ALEXANDER RUDENSKY INTERVENTIONAL IMMUNOLOGIST RADIOLOGIST 16 12 VIVIANE TABAR The clinicians and scientists of NEUROSURGEON Memorial Sloan-Kettering share a vision and 18 a singular mission — to conquer cancer. STEPHEN LONG STRUCTURAL BIOLOGIST They are experts united against a 20 SIMON POWELL complex disease. Each type of cancer R ADIATION ONCOLOGIST 24 ETHEL LAW is different, each tumor is unique. Set free NURSE PRACTITIONER in surroundings that invite the sharing of 26 ideas and resources, they attack the CHRISTINA LESLIE complexity of cancer from every angle COMPUTATIONAL BIOLOGIST and every discipline. 34 SCOTT ARMSTRONG PEDIATRIC ONCOLOGIST 30 TO JORGE REIS-FILHO EXPERIMENTAL PATHOLOGIST CONQUER 38 CANCER 04 Letter from the Chairman and the President A complete version of this report — 42 Statistical Profile which includes lists of our donors, 44 Financial Summary doctors, and scientists — 46 Boards of Overseers and Managers is available on our website at 49 The Campaign for Memorial Sloan-Kettering www.mskcc.org/annualreport. 4 5 Letter from the Chairman In 2012 the leadership of Memorial Sloan-Kettering endorsed Douglas A. Warner III These programmatic investments require leadership and and the President a $2.2 billion investment in a clinical expansion that will set vision. Our new Physician-in-Chief, José Baselga, joined the stage for a changing care paradigm into the next decade us on January 1, 2013. An internationally recognized and beyond. -

The Architecture of a Eukaryotic Replisome
The Architecture of a Eukaryotic Replisome Jingchuan Sun1,2, Yi Shi3, Roxana E. Georgescu3,4, Zuanning Yuan1,2, Brian T. Chait3, Huilin Li*1,2, Michael E. O’Donnell*3,4 1 Biosciences Department, Brookhaven National Laboratory, Upton, New York, USA 2 Department of Biochemistry & Cell Biology, Stony Brook University, Stony Brook, New York, USA. 3 The Rockefeller University, 1230 York Avenue, New York, New York, USA. 4 Howard Hughes Medical Institute *Correspondence and requests for materials should be addressed to M.O.D. ([email protected]) or H.L. ([email protected]) ABSTRACT At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase leads the way in front to unwind DNA, and that DNA polymerases (Pol) trail behind the helicase. Here we use single particle electron microscopy to directly image a replisome. Contrary to expectations, the leading strand Pol ε is positioned ahead of CMG helicase, while Ctf4 and the lagging strand Pol α-primase (Pol α) are behind the helicase. This unexpected architecture indicates that the leading strand DNA travels a long distance before reaching Pol ε, it first threads through the Mcm2-7 ring, then makes a U-turn at the bottom to reach Pol ε at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture, and provides a basis for further structural and biochemical replisome studies. INTRODUCTION DNA is replicated by a multi-protein machinery referred to as a replisome 1,2. Replisomes contain a helicase to unwind DNA, DNA polymerases that synthesize the leading and lagging strands, and a primase that makes short primed sites to initiate DNA synthesis on both strands. -

New Nucleotide Sequence Data on the EMBL File Server
.=) 1991 Oxford University Press Nucleic Acids Research, Vol. 19, No. 2 413 New nucleotide sequence data on the EMBL File Server October 30, 1990 to November 13, 1990 New nucleotide sequence data, available from the EMBL File Server, (see Stoehr, P.J. and Omond, R.A. (1989) Nucleic Acids Res., 17 (16), 6763 -6764), are reported below. Availability of all the newest sequence data is the result of collaboration between.the EMBL Data Library and GenBank' , and data are supplied regularly by both groups. Updates to existing data are not reported here. This report has been prepared by the EMBL Data Library. PRIMATES: Human casein kinase II alpha subunit mRNA, complete cds. Human specific HS5 DNA Lozeman F.J., Litchfield D.W., Piening C., Takio K., Walsh Ueda S., Washio K., Kurosaki K.; K.A., Krebs E.G.; Genomics 8:7-12(1990). X17579 Biochemistry 29:8436-8447(1990). M55268 Human mRNA for heat shock protein HSP27 Human mRNA for actin-binding protein (ABP-280) Carper S.W., Rocheleau T.A., Storm F.K.; Gorlin J.B., Yamin R., Egan S., Stewart M., Stossel T.P., Nucleic Acids Res. 18:6457-6457(1990). X54079 Kwiatkowski D.J., Hartwig J.H.; J. Cell Biol. 111:1089-1105(1990). X53416 Human Ig germline H-chain D-region Dxpl and Dxp'1 genes, 3' Human amiloride-binding protein, complete cds. end. Barbry P., Champe M., Chassande O., Munemitsu S., Champigny Huang C., Stollar B.David.; G., Lingueglia E., Maes P., Frelin C., Tartar A., Ullrich A., Unpublished. M37485 Lazdunski M.; Proc. Natl. Acad. -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Sudips Revised Thesis
Investigation Of The Behavior Of The Gal4 Inhibitor Gal80 Of The GAL Genetic Switch In The Yeast Saccharomyces Cerevisiae Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Sudip Goswami, M.S. Graduate Program in Molecular Genetics The Ohio State University 2014 Dissertation Committee Dr. James Hopper, Advisor Dr. Stephen Osmani Dr. Hay-Oak Park Dr. Jian-Qiu Wu ii Copyright by Sudip Goswami 2014 iii ABSTRACT The DNA-binding transcriptional activator Gal4 and its regulators Gal80 and Gal3 constitute a galactose-responsive switch for the GAL genes of Saccharomyces cerevisiae. Gal4 binds to upstream activation sequences or UASGAL sites on GAL gene promoters as a dimer both in the absence and presence of galactose. In the absence of galactose, a Gal80 dimer binds to and masKs the Gal4 activation domain, inhibiting its activity. In the presence of galactose, Gal3 interacts with Gal80 and relieves Gal80’s inhibition of Gal4 activity allowing rapid induction of expression of GAL genes. In the first part of this work (Chapter 2) in-vitro chemical crosslinking coupled with SDS PAGE and native PAGE analysis were employed to show that the presence of Gal3 that can interact with Gal80 impairs Gal80 self association. In addition, live cell spinning disK confocal imaging showed that dissipation of newly discovered Gal80-2mYFP/2GFP clusters in galactose is dependent on Gal3’s ability to interact with Gal80. In the second part (Chapter 3), extensive analysis of Gal80 clusters was carried out which showed that these clusters associate strongly with the GAL1-10-7 locus and this association is dependent on the presence of the UASGAL sites at this locus.