High Prevalence of Lepus Corsicanus and Hybridization with Introduced L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cahier Des Charges De L'appellation D'origine Contrôlée Vin De Corse Ou
Publié au BO-AGRI le Cahier des charges de l’appellation d’origine contrôlée « VIN DE CORSE » ou « CORSE » homologué par le décret n° 2011-1084 du 8 septembre 2011, modifié par arrêté du publié au JORF du CHAPITRE Ier I. - Nom de l’appellation Seuls peuvent prétendre à l’appellation d’origine contrôlée « Vin de Corse » ou « Corse », initialement reconnue par le décret du 22 décembre 1972, les vins répondant aux dispositions particulières fixées ci- après. II. - Dénominations géographiques et mentions complémentaires 1°- Le nom de l’appellation d’origine contrôlée peut être suivi de la dénomination géographique « Calvi » pour les vins répondant aux conditions de production fixées pour cette dénomination géographique dans le présent cahier des charges. 2°- Le nom de l’appellation d’origine contrôlée peut être suivi de la dénomination géographique « Coteaux du Cap Corse » pour les vins répondant aux conditions de production fixées pour cette dénomination géographique dans le présent cahier des charges. 3°- Le nom de l’appellation d’origine contrôlée peut être suivi de la dénomination géographique « Figari » pour les vins répondant aux conditions de production fixées pour cette dénomination géographique dans le présent cahier des charges. 4°- Le nom de l’appellation d’origine contrôlée peut être suivi de la dénomination géographique « Porto- Vecchio » pour les vins répondant aux conditions de production fixées pour cette dénomination géographique dans le présent cahier des charges. 5°- Le nom de l’appellation d’origine contrôlée peut être suivi de la dénomination géographique « Sartène » pour les vins répondant aux conditions de production fixées pour cette dénomination géographique dans le présent cahier des charges. -

Numéro 012 : Déc.-Janvier 2018 – Nativité
VOIX des clochers de la COSTA VERDE Cervione (St Alexandre et St Erasme) ; Sant’Andria di Cotone (St André, Apôtre) ; Valle di Campoloro (St Augustin ; St Pancrace) ; Santa Maria Poghju (Assunta Gloriosa) ; San Ghjulianu (St Julien et Ste Basilisse) ; San Nicolao (St Nicolas) ; Moriani Plage (N.D. des Flots) ; Santa Lucia di Moriani (Ste Lucie) ; San Ghjuvanni di Moriani (St Jean, Apôtre et Evangéliste) ; Santa Reparata di Moriani (Ste Reparata ; Sant’Antone Abbatu à u porcu) ; Ortale (Assunta Gloriosa) ; Valle d’Alesani (Saints Pierre et Paul) ; Felce (Saints Côme et Damien) ; Tarrano (St Vitus ; Immaculée Conception) ; Piobetta (Annonciation) ; Pietricaggio (Saint Sauveur) ; Perelli ( St Silvestre) ; Piazzali (N .D. d’Alesani ; St Pierre) ; Novale (St Etienne) ; Chiatra di Verde (St Roch ; Annonciation) ; Pietra di Verde (St Elie ; St Augustin ; St Pancrace) BULLETIN INTER-PAROISSIAL www.costaverde.laparoisse.fr 04.95.38.14.84 [email protected] ******************************************************************************************* N°12 : Décembre 2017-Janvier 2018 – Nativité-Epiphanie ******************************************************************************************* Dans ce numéro on trouve : I. NOEL - page 1 II. ANNEE 2017 – Baptêmes ; Communions ; Professions de Foi ; Mariages ; Décès - page 2 III. Programmes des offices / Annonces / Publicité – page 3 ******************************************************************************************* I. NOEL Comme vous le savez tous, la date de mon anniversaire approche. Tous les ans, il y a une grande célébration en mon honneur et je pense que cette année encore cette célébration aura lieu. Pendant cette période, tout le monde fait du shopping, achète des cadeaux, il y a plein de publicité à la radio et dans les magasins, et tout cela augmente au fur et à mesure que mon anniversaire se rapproche. C'est vraiment bien de savoir, qu'au moins une fois par an, certaines personnes pensent à moi. -

Guide Touristique 2020-2021
MARANA-GOLO GUIDE TOURISTIQUE EXPÉRIENCE LA CORSE HORS SECRETS UNIQUE DES SENTIERS BATTUS DE VOYAGE Immersion Escapade Nos favoris en pleine Nature Authentique Parc de Saleccia 03 LES BONNES RAISONS DE VENIR À MARANA-GOLO ! 08 | LA MER 12 | LA NATURE 20 | LA FAMILLE 26 | PARTIR À LA DÉCOUVERTE 50 | NOS TABLES 56 | PRODUCTEURS & ARTISANS 60 | OÙ DORMIR 74 | SE DÉPLACER UN LIEU UNIQUE 05 BIENVENUE À MARANA-GOLO ! Venez passer vos vacances en Corse dans une région unique ! Entité géographique de la plaine orientale de Corse, Marana Golo, ouverte sur la mer Tyrrhénienne, présente les atouts de territoires variés. Face à l’archipel Toscan, de l’embouchure du fleuve du Golo aux portes de la Ville de Bastia, Le Lido de la Marana confine la Réserve Naturelle de l’Étang de Biguglia et intègre un ensemble de microrégions. Entre mer et montagne, Marana-Golo alterne paysages d’exception, sites historiques et villages de caractère aux charmes authentiques, nichés au cœur du piémont, où règnent les senteurs du maquis fait d’arbousiers et de bruyères… Venez vivre une escapade inoubliable… Marana Golo vous attend ! DIRECTION D82 Saint-Florent LÉGENDE 01 La mer . p.09 11 La nature . p.13 D82 14 En famille . p.21 BAIGNADES EN RIVIÈRE D82 26 Accès par Sole e Frescu 30 42.52559, 9.41109 27 Accès par Barchetta 42.50655, 9.37433 28 Accès par le Torrent de Cipetto 42.51023, 9.33905 29 Accès par Accendi pipa 42.50634, 9.31898 30 Accès par le Jardin d’Antoine 42.5933, 9.36345 D5 Bigorno Campitello Scolca Lento D7 D15 28 T20 D5 29 27 DIRECTIONS Corte Île-Rousse -

The State of Lagomorphs Today
HOUSE RABBIT JOURNAL The publication for members of the international House Rabbit Society Winter 2016 The State of Lagomorphs Today by Margo DeMello, PhD Make Mine Chocolate™ Turns 15 by Susan Mangold and Terri Cook Advocating For Rabbits by Iris Klimczuk Fly Strike (Myiasis) in Rabbits by Stacie Grannum, DVM $4.99 CONTENTS HOUSE RABBIT JOURNAL Winter 2016 Contributing Editors Amy Bremers Shana Abé Maureen O’Neill Nancy Montgomery Linda Cook The State of Lagomorphs Today p. 4 Sandi Martin by Margo DeMello, PhD Rebecca Clawson Designer/Editor Sandy Parshall Veterinary Review Linda Siperstein, DVM Executive Director Anne Martin, PhD Board of Directors Marinell Harriman, Founder and Chair Margo DeMello, President Mary Cotter, Vice President Joy Gioia, Treasurer Beth Woolbright, Secretary Dana Krempels Laurie Gigous Kathleen Wilsbach Dawn Sailer Bill Velasquez Judith Pierce Edie Sayeg Nancy Ainsworth House Rabbit Society is a 501c3 and its publication, House Rabbit Journal, is published at 148 Broadway, Richmond, CA 94804. Photograph by Tom Young HRJ is copyright protected and its contents may not be republished without written permission. The Bunny Who Started It All p. 7 by Nareeya Nalivka Goldie is adoptable at House Rabbit Society International Headquarters in Richmond, CA. rabbitcenter.org/adopt Make Mine Chocolate™ Turns 15 p. 8 by Susan Mangold and Terri Cook Cover photo by Sandy Parshall, HRS Program Manager Bella’s Wish p. 9 by Maurice Liang Advocating For Rabbits p. 10 by Iris Klimczuk From Grief to Grace: Maurice, Miss Bean, and Bella p. 12 by Chelsea Eng Fly Strike (Myiasis) in Rabbits p. 13 by Stacie Grannum, DVM The Transpacifi c Bunny p. -

ANTISANTI (Haute Corse)
Dossier Immobilier : ANTISANTI (Haute Corse) Cabinet CORSE EXPERTISE IMMOBILIÈRE & FONCIÈRE Ghjuvan’Santu LE MAO – EXPERT IMMOBILIER AGRÉÉ REV & TRV PAR TEGoVA – CEIF FNAIM – IFEI - EEFIC www.corseexpertiseimmo.com Tél : 04.95.35.00.20 / Mail : [email protected] Siège Social : Avenue du 9 Septembre, Villa Achilli, 20 240 GHISONACCIA Accréditation TEGoVA : REV (Recognised European Valuer) n° REV-FR/CEIF-FNAIM/2020/7 et TRV (TEGoVA Residential Valuer) n° TRV-FR/CEIF-FNAIM/2023/6 Membre de la Chambre des Experts Immobiliers de France FNAIM et de l’Institut Français de l’Expertise Immobilière Membre de la Fédération Nationale des Experts et Experts de Justice Évaluateurs Fonciers, Immobiliers et Commerciaux Contrat R.C.P. MMA IARD ° 120 145 975 R.C Bastia, Siret : 789 004 330 00013 Cabinet CORSE EXPERTISE IMMOBILIERE FONCIERE Ghjuvan’Santu LE MAO - EXPERT IMMOBILIER AGREE REV TRV TEGoVA - CEIF FNAIM - IFEI - EEFIC Évolution de la population sur la Commune (source INSEE) Année Population Densité Population 1968 486 10,10 700 1975 534 11,10 1982 661 13,80 600 1990 500 10,40 500 1999 418 8,70 400 2007 378 7,90 300 2012 429 8,90 200 2017 546 11,40 100 - 1960 1970 1980 1990 2000 2010 2020 Population par grandes tranches d'âges (Source INSEE) 2007 2012 2017 Ensemble 378 100,00% 429 100,00% 546 100,00% 0 à 14 ans 34 8,99% 37 8,62% 65 11,90% 15 à 29 ans 52 13,76% 50 11,66% 42 7,69% 30 à 44 ans 53 14,02% 68 15,85% 112 20,51% 45 à 59 ans 83 21,96% 94 21,91% 107 19,60% 60 à 74 ans 99 26,19% 109 25,41% 135 24,73% 75 ans ou 57 15,08% 71 16,55% -

Dossier EPCI
N° 11 Octobre 2018 Portrait des 19 intercommunalités de Corse Synthèse Sommaire Synthèse Dans le cadre de la loi sur la Nouvelle Organisation Territoriale de la Les 19 intercommunalités de corse République (NOTRe), une nouvelle cartographie des intercommunalités est effective depuis le 1er janvier 2017. Sur l'ensemble du territoire de Corse, cela se traduit par la création de 19 établissements publics de coopération intercommunale (EPCI) : Synthèse 3 deux communautés d'agglomération (CA) et 17 communautés de Cap communes (CC). Cette refonte réduit le nombre d'EPCI qui était de Corse 27 auparavant ; huit intercommunalités ont gardé leurs délimitations Communauté d'agglomération du Pays Ajaccien 4 antérieures. Ces modifications sont principalement dues à la taille minimum demandée par la loi : 15 000 habitants, avec une Communauté d'agglomération de Bastia 7 Nebbiu- Bastia dérogation à 5 000 pour les territoires montagneux. La Corse étant Conca d'Oro une « montagne dans la mer », seuls quatre EPCI dépassent les Communauté de communes de Marana-Golo 10 L'Île-Rousse-Balagne 15 000 résidents : les deux CA du Pays Ajaccien et de Bastia ainsi Marana-Golo Communauté de communes du Sud Corse que les CC de Marana-Golo et du Sud Corse. 13 Castagniccia- Communauté de communes de Fium'orbu Castellu Des territoires aux profils différents Calvi Balagne Casinca 16 Pasquale Paoli Communauté de communes de la Castagniccia-Casinca Les intercommunalités de l'île sont plurielles. Avec 917 km², la CC 19 Costa Verde Communauté de communes de la Pieve de l'Ornano Spelunca-Liamone est la plus vaste, 13 fois plus étendue que la plus 22 petite, la CA de Bastia. -

17 Décembre 2019 Page 1 CONVENTION QUINQUENNALE 2020-2024 PREAMBULE La Révision De La Charte Du SM/PNRC : D'une Période D
CONVENTION QUINQUENNALE 2020-2024 RELATIVE A LA DEFINITION ET A LA MISE EN ŒUVRE DES ACTIONS DU SYNDICAT MIXTE DU PARC NATUREL REGIONAL DE CORSE - PARCU DI CORSICA SUR SON TERRITOIRE PREAMBULE La révision de la Charte du SM/PNRC : d’une période d’incertitude à une concertation élargie et déterminante : Le classement du Parc Naturel Régional de Corse (PNRC) avait été renouvelé pour 10 ans par décret du 9 juin 1999 sur un territoire de 145 communes, étendu à deux communes supplémentaires par décret du 12 avril 2007. Une première révision de la charte du Parc Naturel Régional de Corse, avait été prescrite par délibération de l’Assemblée de Corse en date du 30 mars 2007. Elle était initialement envisagée sur un périmètre d’étude identique à celui du périmètre classé en Parc naturel régional. Un débat s’est ensuite instauré sur l’opportunité d’étendre ce périmètre, certains élus considérant que la valeur patrimoniale de la Corse justifiait l’inscription de l’ensemble de l’île en Parc naturel régional. Le diagnostic territorial réalisé en 2011 à la demande de la Collectivité Territoriale de Corse (CTC) a permis d’analyser les possibilités d’extensions pertinentes au regard des critères de classement d’un Parc naturel régional, tels qu’ils sont définis par le code de l’environnement. Le classement du Parc a été prolongé par décret du 2 juin 2009 jusqu’au 9 juin 2011. Depuis cette date, le Parc Naturel Régional de Corse n’était plus classé. La révision de la Charte a par la suite été relancée en juillet 2013, selon un processus concerté avec l’Office de l’Environnement de la Corse, la fédération des parcs naturels régionaux de France et en relation étroite avec l’Etat. -

Liste Des Électeurs Sénatoriaux En Haute-Corse
PRÉFECTURE DE LA HAUTE-CORSE ÉLECTIONS SÉNATORIALES du 27 septembre 2020 ********** Liste des électeurs sénatoriaux (dressé en application des dispositions de l'article R. 146 du code électoral) 1 / 49 1 – SÉNATEUR - CASTELLI Joseph 2 – DÉPUTÉS - ACQUAVIVA Jean-Félix - CASTELLANI Michel 3 - CONSEILLERS A L’ASSEMBLÉE DE CORSE - ARMANET Guy - ARRIGHI Véronique - BENEDETTI François - CARLOTTI Pascal - CASANOVA-SERVAS Marie-Hélène - CECCOLI François-Xavier - CESARI Marcel - COGNETTI-TURCHINI Catherine - DELPOUX Jean-Louis - DENSARI Frédérique - GHIONGA Pierre - GIUDICI Francis - GIOVANNINI Fabienne - GRIMALDI Stéphanie - MARIOTTI Marie-Thérèse - MONDOLONI Jean-Martin - MOSCA Paola - NIVAGGIONI Nadine - ORLANDI François - PADOVANI Marie-Hélène - PAOLINI Julien - PARIGI Paulu-Santu - PIERI Marie-Anne - POLI Antoine - PONZEVERA Juliette - POZZO di BORGO Louis - PROSPERI Rosa - SANTUCCI Anne-Laure - SIMEONI Marie - SIMONI Pascale - TALAMONI Jean-Guy - TOMASI Anne - TOMASI Petr’Antone - VANNI Hyacinthe 4 - DÉLÉGUÉS DES CONSEILS MUNICIPAUX -AGHIONE a) Délégué élu - CASANOVA André b) Suppléants - VIELES Francis - BARCELO Daniel - KLEINEIDAM Hervé -AITI a) Délégué élu - ORSONI Gérard b) Suppléants - ANGELI Marius -ALANDO a) Délégué élu - MAMELLI Guy b) Suppléants - IMPERINETTI Jean-Raphaël - POLETTI Cécile - BERNARDI Jean-François Albert -ALBERTACCE a) Délégué élu - ALBERTINI Pierre-François b) Suppléants - SANTINI Pasquin - ALBERTINI Catherine - ZAMBARO Flavia -ALERIA a) Délégués élus - LUCIANI Dominique - TADDEI Laurence - CHEYNET Patrick - HERMÉ -

Avis De La Mission Régionale D'autorité Environnementale De
Corse Avis de la Mission Régionale d’Autorité environnementale de Corse sur l’élaboration du plan local d’urbanisme de VICO (Corse-du-Sud) n°MRAe 2018-02 1 AVIS DÉLIBÉRÉ N° 2018-02 adopté lors de la séance du 11 juin 2018 par La mission régionale d’autorité environnementale de Corse Préambule La Mission régionale d’autorité environnementale (MRAe) de Corse s’est réunie téléphoniquement le XXXXX 2018. L’ordre du jour comportait notamment, l’avis sur l’élaboration du plan local d’urbanisme (PLU) de la commune de Vico. Étaient présents et ont délibéré : Fabienne Allag-Dhuisme présidente, Jet en tant que membres associés, Marie Livia Leoni et Louis Olivier ; Etait excusé : Jean-Pierre Viguier, membre permanent titulaire. Était présent sans voix délibérative : Jean-Marie Seité membre associé suppléant. En application de l’article 9 du règlement intérieur du CGEDD, chacun des membres délibérants cités ci- dessus atteste qu’aucun intérêt particulier ou élément dans ses activités passées ou présentes n’est de nature à mettre en cause son impartialité dans l’avis à donner sur le projet qui fait l’objet du présent avis. L’ordonnance n°2004-489 du 3 juin 2004, portant transposition de la directive 2001/42/CE du parlement européen et du Conseil du 27 juin 2001, a introduit la notion d’évaluation des incidences de certains plans et programmes sur l’environnement. Le décret n°2005-608 du 27 mai 2005 a complété le code de l’urbanisme par les articles désormais codifiés R. 104-1 et suivants. La procédure d’évaluation environnementale, diligentée au stade de la planification, en amont des projets opérationnels, vise à repérer de façon préventive les impacts potentiels des grandes orientations du document d’urbanisme sur l’environnement, à un stade où les infléchissements sont plus aisés à mettre en œuvre. -
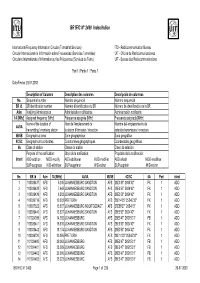
BR IFIC N° 2499 Index/Indice
BR IFIC N° 2499 Index/Indice International Frequency Information Circular (Terrestrial Services) ITU - Radiocommunication Bureau Circular Internacional de Información sobre Frecuencias (Servicios Terrenales) UIT - Oficina de Radiocomunicaciones Circulaire Internationale d'Information sur les Fréquences (Services de Terre) UIT - Bureau des Radiocommunications Part 1 / Partie 1 / Parte 1 Date/Fecha: 29.07.2003 Description of Columns Description des colonnes Descripción de columnas No. Sequential number Numéro séquenciel Número sequencial BR Id. BR identification number Numéro d'identification du BR Número de identificación de la BR Adm Notifying Administration Administration notificatrice Administración notificante 1A [MHz] Assigned frequency [MHz] Fréquence assignée [MHz] Frecuencia asignada [MHz] Name of the location of Nom de l'emplacement de Nombre del emplazamiento de 4A/5A transmitting / receiving station la station d'émission / réception estación transmisora / receptora 4B/5B Geographical area Zone géographique Zona geográfica 4C/5C Geographical coordinates Coordonnées géographiques Coordenadas geográficas 6A Class of station Classe de station Clase de estación Purpose of the notification: Objet de la notification: Propósito de la notificación: Intent ADD-addition MOD-modify ADD-additioner MOD-modifier ADD-añadir MOD-modificar SUP-suppress W/D-withdraw SUP-supprimer W/D-retirer SUP-suprimir W/D-retirar No. BR Id Adm 1A [MHz] 4A/5A 4B/5B 4C/5C 6A Part Intent 1 100039437 AFS 5.245 JOHANNESBURG SANDTON AFS 28E3'53" 26S4'42" FX 1 ADD -

Appel À Candidatures Du 03 Janv Au 21 Janv 2019
Publication effectuée en application des articles L 141-1, L 141-3, L 143-3 et R 142-3 du Code Rural et de la Pêche Maritime La SAFER CORSE se propose, sans engagement de sa part, d’attribuer par RETROCESSION, SUBSTITUTION ou LOCATION, tous ou parties des Biens désignés ci-après qu’elle possède ou qu’elle envisage d’acquérir. Les propriétés ci-après désignées sont mises en attributions sous réserve de la bonne finalité des opérations d'acquisition par la SAFER Corse. 1°) RETROCESSION-SUBSTITUTION ANTISANTI : 1 ha 18 a 40 ca : Lieudit Maccione : Section ZV n°40 En nature : Terre et maquis - (sans bâtiment) Classification dans un document d'urbanisme : R.N.U. (Règlement National d’Urbanisme) ANTISANTI : 18 ha 79 a 84 ca : Lieudit Agno di pino : Section ZR-16p-20p– Lieudit Fontana calda : Section E n°320-321-322-326-327 – Lieudit Limeruccia : Section A n°10-11 – Lieudit Linarello : Section ZW-7p – Lieudit Piglia pecora : Section YA n° 41 – Lieudit San margini : Section A n°18 En nature : Friches et maquis. (sans bâtiment) Classification dans un document d'urbanisme : R.N.U. (Règlement National d’Urbanisme) ANTISANTI : 15 ha 92 a 00 ca : Lieudit Pianiccia olivella : Section ZH n° 5-31-32 En nature : Vigne, vergers, friches et maquis (sans bâtiment) Classification dans un document d'urbanisme : R.N.U. (Règlement National d’Urbanisme) BARBAGGIO : 40 a 75 ca : Lieudit Calarana : Section A n° 848- Lieudit Pian del casale : Lieudit A n° 540-542- 1042-1044 En nature : Bois et verger Bâtis : Maison d'habitation de type 4 d’environ 90 m² Piscine -

World Distribution of the European Rabbit (Oryctolagus Cuniculus)
1 The Evolution, Domestication and World Distribution of the European Rabbit (Oryctolagus cuniculus) Luca Fontanesi1*, Valerio Joe Utzeri1 and Anisa Ribani1 1Department of Agricultural and Food Sciences, Division of Animal Sciences, University of Bologna, Italy 1.1 The Order Lagomorpha to assure essential vitamin uptake, the digestion of the vegetarian diet and water reintroduction The European rabbit (Oryctolagus cuniculus, (Hörnicke, 1981). Linnaeus 1758) is a mammal belonging to the The order Lagomorpha was recognized as a order Lagomorpha. distinct order within the class Mammalia in Lagomorphs are such a distinct group of 1912, separated from the order Rodentia within mammalian herbivores that the very word ‘lago- which lagomorphs were originally placed (Gidely, morph’ is a circular reference meaning ‘hare- 1912; Landry, 1999). Lagomorphs are, however, shaped’ (Chapman and Flux, 1990; Fontanesi considered to be closely related to the rodents et al., 2016). A unique anatomical feature that from which they diverged about 62–100 million characterizes lagomorphs is the presence of years ago (Mya), and together they constitute small peg-like teeth immediately behind the up- the clade Glires (Chuan-Kuei et al., 1987; Benton per-front incisors. For this feature, lagomorphs and Donoghue, 2007). Lagomorphs, rodents and are also known as Duplicidentata. Therefore, primates are placed in the major mammalian instead of four incisor teeth characteristic of clade of the Euarchontoglires (O’Leary et al., 2013). rodents (also known as Simplicidentata), lago- Modern lagomorphs might be evolved from morphs have six. The additional pair is reduced the ancestral lineage from which derived the in size. Another anatomical characteristic of the †Mimotonidae and †Eurymilydae sister taxa, animals of this order is the presence of an elong- following the Cretaceous-Paleogene (K-Pg) bound- ated rostrum of the skull, reinforced by a lattice- ary around 65 Mya (Averianov, 1994; Meng et al., work of bone, which is a fenestration to reduce 2003; Asher et al., 2005; López-Martínez, 2008).