Survey of Leptospira Spp in Pampas Deer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Redalyc.ECOLOGICAL STRATEGIES and IMPACT of WILD BOAR IN
Mastozoología Neotropical ISSN: 0327-9383 [email protected] Sociedad Argentina para el Estudio de los Mamíferos Argentina Cuevas, M. Fernanda; Ojeda, Ricardo A.; Jaksic, Fabian M. ECOLOGICAL STRATEGIES AND IMPACT OF WILD BOAR IN PHYTOGEOGRAPHIC PROVINCES OF ARGENTINA WITH EMPHASIS ON ARIDLANDS Mastozoología Neotropical, vol. 23, núm. 2, 2016, pp. 239-254 Sociedad Argentina para el Estudio de los Mamíferos Tucumán, Argentina Available in: http://www.redalyc.org/articulo.oa?id=45750282004 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Mastozoología Neotropical, 23:239-254, Mendoza, 2016 Copyright ©SAREM, 2016 http://www.sarem.org.ar Versión impresa ISSN 0327-9383 http://www.sbmz.com.br Versión on-line ISSN 1666-0536 Sección Especial MAMÍFEROS EXÓTICOS INVASORES ECOLOGICAL STRATEGIES AND IMPACT OF WILD BOAR IN PHYTOGEOGRAPHIC PROVINCES OF ARGENTINA WITH EMPHASIS ON ARIDLANDS M. Fernanda Cuevas1, Ricardo A. Ojeda1 and Fabian M. Jaksic2 1 Grupo de Investigaciones de la Biodiversidad (GiB), IADIZA, CCT Mendoza CONICET, CC 507, 5500 Mendoza, Argentina. [Correspondence: <[email protected]>]. 2 Centro de Ecología Aplicada y Sustentabilidad (CAPES), Pontificia Universidad Católica de Chile, Casilla 114-D, Santiago, Chile ABSTRACT. Wild boar is an invasive species introduced to Argentina for sport hunting purposes. Here, this spe- cies is present in at least 8 phytogeographic provinces but we only have information in four of them (Pampean grassland, Espinal, Subantarctic and Monte Desert). We review the ecological strategies and impact of wild boar on ecosystem processes in these different phytogeographic provinces and identify knowledge gaps and research priorities for a better understanding of this invasive species in Argentina. -

Reproductive Seasonality in Captive Wild Ruminants: Implications for Biogeographical Adaptation, Photoperiodic Control, and Life History
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2012 Reproductive seasonality in captive wild ruminants: implications for biogeographical adaptation, photoperiodic control, and life history Zerbe, Philipp Abstract: Zur quantitativen Beschreibung der Reproduktionsmuster wurden Daten von 110 Wildwiederkäuer- arten aus Zoos der gemässigten Zone verwendet (dabei wurde die Anzahl Tage, an denen 80% aller Geburten stattfanden, als Geburtenpeak-Breite [BPB] definiert). Diese Muster wurden mit verschiede- nen biologischen Charakteristika verknüpft und mit denen von freilebenden Tieren verglichen. Der Bre- itengrad des natürlichen Verbreitungsgebietes korreliert stark mit dem in Menschenobhut beobachteten BPB. Nur 11% der Spezies wechselten ihr reproduktives Muster zwischen Wildnis und Gefangenschaft, wobei für saisonale Spezies die errechnete Tageslichtlänge zum Zeitpunkt der Konzeption für freilebende und in Menschenobhut gehaltene Populationen gleich war. Reproduktive Saisonalität erklärt zusätzliche Varianzen im Verhältnis von Körpergewicht und Tragzeit, wobei saisonalere Spezies für ihr Körpergewicht eine kürzere Tragzeit aufweisen. Rückschliessend ist festzuhalten, dass Photoperiodik, speziell die abso- lute Tageslichtlänge, genetisch fixierter Auslöser für die Fortpflanzung ist, und dass die Plastizität der Tragzeit unterstützend auf die erfolgreiche Verbreitung der Wiederkäuer in höheren Breitengraden wirkte. A dataset on 110 wild ruminant species kept in captivity in temperate-zone zoos was used to describe their reproductive patterns quantitatively (determining the birth peak breadth BPB as the number of days in which 80% of all births occur); then this pattern was linked to various biological characteristics, and compared with free-ranging animals. Globally, latitude of natural origin highly correlates with BPB observed in captivity, with species being more seasonal originating from higher latitudes. -
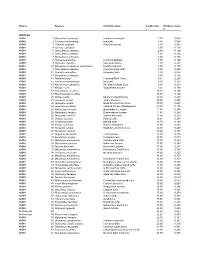
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

Behavioral Responses of the Pampas Deer (Ozotoceros Bezoarticus) to Human Disturbance in San Luis Province, Argentina
NORTH-WESTERN JOURNAL OF ZOOLOGY 13 (1): 159-162 ©NwjZ, Oradea, Romania, 2017 Article No.: e162701 http://biozoojournals.ro/nwjz/index.html Behavioral responses of the pampas deer (Ozotoceros bezoarticus) to human disturbance in San Luis province, Argentina María Belén SEMEÑIUK1,2,* and Mariano Lisandro MERINO3,4 1. Anexo Museo de La Plata, Universidad Nacional de La Plata, 122 y 60, La Plata, 1900, Buenos Aires, Argentina. 2. Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). 3. Centro de Bioinvestigaciones, Universidad Nacional del Noroeste de la Provincia de Buenos Aires, CIT-NOBA, Ruta Provincial 32 Km 3.5, Pergamino 2700, Buenos Aires, Argentina. 4. Comisión de Investigaciones Científicas de la provincia de Buenos Aires. *Corresponding author, M.B. Semeñiuk, Email: [email protected] Received: 05. October 2015 / Accepted: 14. March 2016 / Available online: 26. June 2016 / Printed: June 2017 Abstract. The pampas deer Ozotoceros bezoarticus (Linnaeus, 1758) is a South American cervid, associated with grasslands and savannas; in Argentina this species is listed as “endangered”. Our aim was to analyze the behavioral responses of the pampas deer to human presence, and to evaluate possible effects of their poaching. We recorded behavioral responses from 382 pampas deer groups during eight vehicle surveys, in “El Centenario” ranch (San Luis Province). Data were analyzed using the G-test of independence and logistic regression. Almost half of the groups (48.17 percent) remained on site. Behavioral responses differed significantly according to group size and composition and observer distance, with the last variable having the greatest influence on flight - groups were more likely to flee at shorter distances. -

Sexual Selection and Extinction in Deer Saloume Bazyan
Sexual selection and extinction in deer Saloume Bazyan Degree project in biology, Master of science (2 years), 2013 Examensarbete i biologi 30 hp till masterexamen, 2013 Biology Education Centre and Ecology and Genetics, Uppsala University Supervisor: Jacob Höglund External opponent: Masahito Tsuboi Content Abstract..............................................................................................................................................II Introduction..........................................................................................................................................1 Sexual selection........................................................................................................................1 − Male-male competition...................................................................................................2 − Female choice.................................................................................................................2 − Sexual conflict.................................................................................................................3 Secondary sexual trait and mating system. .............................................................................3 Intensity of sexual selection......................................................................................................5 Goal and scope.....................................................................................................................................6 Methods................................................................................................................................................8 -

Endangered Species (Import and Export) Act (Chapter 92A)
1 S 23/2005 First published in the Government Gazette, Electronic Edition, on 11th January 2005 at 5:00 pm. NO.S 23 ENDANGERED SPECIES (IMPORT AND EXPORT) ACT (CHAPTER 92A) ENDANGERED SPECIES (IMPORT AND EXPORT) ACT (AMENDMENT OF FIRST, SECOND AND THIRD SCHEDULES) NOTIFICATION 2005 In exercise of the powers conferred by section 23 of the Endangered Species (Import and Export) Act, the Minister for National Development hereby makes the following Notification: Citation and commencement 1. This Notification may be cited as the Endangered Species (Import and Export) Act (Amendment of First, Second and Third Schedules) Notification 2005 and shall come into operation on 12th January 2005. Deletion and substitution of First, Second and Third Schedules 2. The First, Second and Third Schedules to the Endangered Species (Import and Export) Act are deleted and the following Schedules substituted therefor: ‘‘FIRST SCHEDULE S 23/2005 Section 2 (1) SCHEDULED ANIMALS PART I SPECIES LISTED IN APPENDIX I AND II OF CITES In this Schedule, species of an order, family, sub-family or genus means all the species of that order, family, sub-family or genus. First column Second column Third column Common name for information only CHORDATA MAMMALIA MONOTREMATA 2 Tachyglossidae Zaglossus spp. New Guinea Long-nosed Spiny Anteaters DASYUROMORPHIA Dasyuridae Sminthopsis longicaudata Long-tailed Dunnart or Long-tailed Sminthopsis Sminthopsis psammophila Sandhill Dunnart or Sandhill Sminthopsis Thylacinidae Thylacinus cynocephalus Thylacine or Tasmanian Wolf PERAMELEMORPHIA -

List of 28 Orders, 129 Families, 598 Genera and 1121 Species in Mammal Images Library 31 December 2013
What the American Society of Mammalogists has in the images library LIST OF 28 ORDERS, 129 FAMILIES, 598 GENERA AND 1121 SPECIES IN MAMMAL IMAGES LIBRARY 31 DECEMBER 2013 AFROSORICIDA (5 genera, 5 species) – golden moles and tenrecs CHRYSOCHLORIDAE - golden moles Chrysospalax villosus - Rough-haired Golden Mole TENRECIDAE - tenrecs 1. Echinops telfairi - Lesser Hedgehog Tenrec 2. Hemicentetes semispinosus – Lowland Streaked Tenrec 3. Microgale dobsoni - Dobson’s Shrew Tenrec 4. Tenrec ecaudatus – Tailless Tenrec ARTIODACTYLA (83 genera, 142 species) – paraxonic (mostly even-toed) ungulates ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BOVIDAE (46 genera) - cattle, sheep, goats, and antelopes 1. Addax nasomaculatus - Addax 2. Aepyceros melampus - Impala 3. Alcelaphus buselaphus - Hartebeest 4. Alcelaphus caama – Red Hartebeest 5. Ammotragus lervia - Barbary Sheep 6. Antidorcas marsupialis - Springbok 7. Antilope cervicapra – Blackbuck 8. Beatragus hunter – Hunter’s Hartebeest 9. Bison bison - American Bison 10. Bison bonasus - European Bison 11. Bos frontalis - Gaur 12. Bos javanicus - Banteng 13. Bos taurus -Auroch 14. Boselaphus tragocamelus - Nilgai 15. Bubalus bubalis - Water Buffalo 16. Bubalus depressicornis - Anoa 17. Bubalus quarlesi - Mountain Anoa 18. Budorcas taxicolor - Takin 19. Capra caucasica - Tur 20. Capra falconeri - Markhor 21. Capra hircus - Goat 22. Capra nubiana – Nubian Ibex 23. Capra pyrenaica – Spanish Ibex 24. Capricornis crispus – Japanese Serow 25. Cephalophus jentinki - Jentink's Duiker 26. Cephalophus natalensis – Red Duiker 1 What the American Society of Mammalogists has in the images library 27. Cephalophus niger – Black Duiker 28. Cephalophus rufilatus – Red-flanked Duiker 29. Cephalophus silvicultor - Yellow-backed Duiker 30. Cephalophus zebra - Zebra Duiker 31. Connochaetes gnou - Black Wildebeest 32. Connochaetes taurinus - Blue Wildebeest 33. Damaliscus korrigum – Topi 34. -

Biological and Social Issues Related to Confinement of Wild Ungulates
Biological and Social Issues Related to Confinement of Wild Ungulates THE WILDLIFE SOCIETY Technical Review 02-3 2002 BIOLOGICALANDSOCIALISSUESRELATEDTOCONFINEMENTOFWILDUNGULATES The Wildlife Society Members of The Technical Committee on Wild Ungulate Confinement Stephen Demarais (Chair) Elizabeth S. Williams Department of Wildlife and Fisheries Department of Veterinary Sciences Mississippi State University University of Wyoming Box 9690 1174 Snowy Range Road Mississippi State, MS 39762 Laramie, WY 82070 Randall W. DeYoung Scot J. Williamson Department of Wildlife and Fisheries Wildlife Management Institute Mississippi State University RR 1, Box 587, Spur Road Box 9690 North Stratford, NH 03590 Mississippi State, MS 39762 Gary J. Wolfe L. Jack Lyon Rocky Mountain Elk Foundation Box 9045 4722 Aspen Drive Missoula, MT 59807 Missoula, MT 59802 Edited by Laura Andrews The Wildlife Society Technical Review 02-3 5410 Grosvenor Lane, Suite 200 November 2002 Bethesda, Maryland 20814 Foreword Presidents of The Wildlife Society occasionally appoint ad hoc committees to study and report on selected conservation issues. The reports ordinarily appear in 2 related series called either Technical Review (formerly "White Paper") or Position Statement. Review papers present technical information and the views of the appointed committee members, but not necessarily the views of their employers. Position statements are based on the review papers, and the preliminary versions ordinarily are published in The Wildliferfor comment by Society members. Following the comment period, revision, and Council's approval, the statements are published as official positions of The Wildlife Society. Both types of reports are copyrighted by the Society, but individuals are granted permission to make single copies for noncommercial purposes. -

Gross Anatomy of the Intestine in the Giraffe (Giraffa Camelopardalis)
Pérez, W; Lima, M; Clauss, M (2009). Gross Anatomy of the Intestine in the Giraffe (Giraffa camelopardalis). Anatomia, Histologia, Embryologia: Journal of Veterinary Medicine Series C, 38(6):432-435. Postprint available at: http://www.zora.uzh.ch University of Zurich Posted at the Zurich Open Repository and Archive, University of Zurich. Zurich Open Repository and Archive http://www.zora.uzh.ch Originally published at: Anatomia, Histologia, Embryologia: Journal of Veterinary Medicine Series C 2009, 38(6):432-435. Winterthurerstr. 190 CH-8057 Zurich http://www.zora.uzh.ch Year: 2009 Gross Anatomy of the Intestine in the Giraffe (Giraffa camelopardalis) Pérez, W; Lima, M; Clauss, M Pérez, W; Lima, M; Clauss, M (2009). Gross Anatomy of the Intestine in the Giraffe (Giraffa camelopardalis). Anatomia, Histologia, Embryologia: Journal of Veterinary Medicine Series C, 38(6):432-435. Postprint available at: http://www.zora.uzh.ch Posted at the Zurich Open Repository and Archive, University of Zurich. http://www.zora.uzh.ch Originally published at: Anatomia, Histologia, Embryologia: Journal of Veterinary Medicine Series C 2009, 38(6):432-435. Gross Anatomy of the Intestine in the Giraffe (Giraffa camelopardalis) Abstract We describe the macroscopic anatomy of the intestine of the giraffe (Giraffa camelopardalis). The small intestine was divided into duodenum, jejunum and ileum as usual. The caecum was attached to the ileum by a long ileocaecal fold, and to the proximal ansa of the ascending colon by a caecocolic fold. The ascending colon was the most developed portion of the gross intestine and had the most complex arrangement with three ansae: the proximal ansa, the spiral ansa and the distal ansa. -

New Populations of Pampas Deer Ozotoceros Bezoarticus Discovered in Threatened Amazonian Savannah Enclaves
New populations of pampas deer Ozotoceros bezoarticus discovered in threatened Amazonian savannah enclaves D ANIEL G. ROCHA,ALEXANDRE V OGLIOTTI,DIOGO M. GRÄBIN W ILHAN R. C. ASSUNÇÃO,BRUNO C ONTURSI C AMBRAIA,ANA R AFAELA D ’ A MICO A NTONIO E LSON P ORTELA and R AHEL S OLLMANN Abstract The savannah enclaves (i.e. patches) in the south- Supplementary material for this article is available at ern Brazilian Amazonia are among the most threatened and https://doi.org/./S poorly surveyed sites in Amazonia. As part of an extensive mammal survey, we set camera traps in three of these savannah enclaves. We obtained independent records he South American savannah is the largest and most of pampas deer Ozotoceros bezoarticus, a medium sized Tbiodiverse tropical savannah (Cardoso Da Silva & Bates, Neotropical cervid that is strongly associated with open ). Most of this biome, known as cerrado, is located in habitats and categorized as Vulnerable on the Brazilian Red central Brazil, and is categorized as one of global conserva- List of threatened species. These savannah enclaves with tion hotspots (Myers et al., ). More than % of the cer- confirmed populations of pampas deer lie outside the rado has been cleared and only .% lies within protected species’ previously presumed historical range and are at areas (Strassburg et al., ). Significant patches of savannah, least km from any known extant population. Together, known as enclaves, can also be found on the periphery of the these savannah enclaves add c. , km to the pampas Amazon forest. These enclaves are heterogeneous mosaics of deer’s currently known range. -

Fertility Reduction in Male Persian Fallow Deer (Dama Dama Mesopotamica): Inbreeding Detection and Morphometric Parameters Evaluation of Semen
Journal of Biosciences and Medicines, 2016, 4, 31-38 Published Online June 2016 in SciRes. http://www.scirp.org/journal/jbm http://dx.doi.org/10.4236/jbm.2016.46005 Fertility Reduction in Male Persian Fallow Deer (Dama dama mesopotamica): Inbreeding Detection and Morphometric Parameters Evaluation of Semen B. Ekrami1*, A. Tamadon2*, I. Razeghian Jahromi2, D. Moghadas3, M. M. Ghahramani Seno4, M. Ghaderi-Zefrehei5 1Department of Obstetrics and Animal Reproductive Diseases, Islamic Azad University of Chalus, College of Agriculture and Livestock Sciences, Chalus & Veterinarian of Environment Department of Mazandaran Province, Sari, Iran 2Transgenic Technology Research Center, Shiraz University of Medical Sciences, Shiraz, Iran 3Department of Environment of Mazandaran province, Sari, Iran 4Department of Basic Sciences, Faculty of Veterinary Medicine, Ferdowsi University of Mashhad, Mashhad, Iran 5Department of Animal Sciences, Faculty of Agriculture, University of Yasouj, Yasouj, Iran Received 8 April 2016; accepted 21 May 2016; published 24 May 2016 Abstract Persian fallow deer (Dama dama mesopotamica) is only found in a few protected and refuges areas in the northwest, north, and southwest of Iran. The aims of this study were analysis of in- breeding and morphometric parameters of semen in male Persian fallow deer to investigate the cause of reduced fertility of this endangered species in Dasht-e-Naz National Refuge, Sari, Iran. The Persian fallow deer semen was collected by an electroejaculator from four adult bucks randomly during the breeding season and from five dehorned and horned deer’s in non-breeding season. Twelve blood samples were taken and mitochondrial DNA was extracted, a non-coding region called d-loop was amplified, sequenced and then were considered for genetic analysis. -

List of Taxa for Which MIL Has Images
LIST OF 27 ORDERS, 163 FAMILIES, 887 GENERA, AND 2064 SPECIES IN MAMMAL IMAGES LIBRARY 31 JULY 2021 AFROSORICIDA (9 genera, 12 species) CHRYSOCHLORIDAE - golden moles 1. Amblysomus hottentotus - Hottentot Golden Mole 2. Chrysospalax villosus - Rough-haired Golden Mole 3. Eremitalpa granti - Grant’s Golden Mole TENRECIDAE - tenrecs 1. Echinops telfairi - Lesser Hedgehog Tenrec 2. Hemicentetes semispinosus - Lowland Streaked Tenrec 3. Microgale cf. longicaudata - Lesser Long-tailed Shrew Tenrec 4. Microgale cowani - Cowan’s Shrew Tenrec 5. Microgale mergulus - Web-footed Tenrec 6. Nesogale cf. talazaci - Talazac’s Shrew Tenrec 7. Nesogale dobsoni - Dobson’s Shrew Tenrec 8. Setifer setosus - Greater Hedgehog Tenrec 9. Tenrec ecaudatus - Tailless Tenrec ARTIODACTYLA (127 genera, 308 species) ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BALAENIDAE - bowheads and right whales 1. Balaena mysticetus – Bowhead Whale 2. Eubalaena australis - Southern Right Whale 3. Eubalaena glacialis – North Atlantic Right Whale 4. Eubalaena japonica - North Pacific Right Whale BALAENOPTERIDAE -rorqual whales 1. Balaenoptera acutorostrata – Common Minke Whale 2. Balaenoptera borealis - Sei Whale 3. Balaenoptera brydei – Bryde’s Whale 4. Balaenoptera musculus - Blue Whale 5. Balaenoptera physalus - Fin Whale 6. Balaenoptera ricei - Rice’s Whale 7. Eschrichtius robustus - Gray Whale 8. Megaptera novaeangliae - Humpback Whale BOVIDAE (54 genera) - cattle, sheep, goats, and antelopes 1. Addax nasomaculatus - Addax 2. Aepyceros melampus - Common Impala 3. Aepyceros petersi - Black-faced Impala 4. Alcelaphus caama - Red Hartebeest 5. Alcelaphus cokii - Kongoni (Coke’s Hartebeest) 6. Alcelaphus lelwel - Lelwel Hartebeest 7. Alcelaphus swaynei - Swayne’s Hartebeest 8. Ammelaphus australis - Southern Lesser Kudu 9. Ammelaphus imberbis - Northern Lesser Kudu 10. Ammodorcas clarkei - Dibatag 11. Ammotragus lervia - Aoudad (Barbary Sheep) 12.