Neurodevelopment Next-Generation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Inherited Metabolic Disorders News
The Inherited Metabolic Disorders News Summer 2011 Volume 8 Issue 2 From the Editor I hope everyone is enjoying their summer thus far! Our 8th annual Metabolic Family Day and 7th annual Low In this Issue Protein Cooking Demonstration were once again a huge success. See the section ”What’s New” on page 9 for a full report of the events, as well as pictures. ♦ From the Editor…1 As always, your suggestions and stories are welcome. Please contact me by email: [email protected] or telephone 519-685-8453 if you wish to contribute to the ♦ From Dr. Chitra Prasad…1 newsletter. I hope everyone has a safe and happy summer! ♦ Personal Stories… 2 Janice Little ♦ Featured This Issue … 4 From Dr Chitra Prasad ♦ Suzanne’s Corner… 6 Dear Friends, ♦ What’s New… 8 Hope you all are having a wonderful summer. We had a great metabolic family workshop with around 198 registrants this year. This was truly phenomenal. Thanks to all the team members and ♦ Research & Presentations … 13 the families in the planning committee for doing such a fantastic job. I was really touched when one of our young metabolic patients told her parents that she would like to attend the ♦ How to Make a Donation… 14 metabolic family workshop! Please see some of the highlights of the metabolic workshop in the newsletter for those who could not make it. The speeches by our youth (Sadiq, Leanna and Laura) ♦ Contact Information … 15 were appreciated by everyone. Big thanks to Jill Tosswill (our previous social worker) who coordinated their talks. -

Moderate the MAOA-L Allele Expression with CRISPR/Cas9 System
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 23 April 2018 doi:10.20944/preprints201804.0275.v1 1 Review 2 Moderate the MAOA-L Allele Expression with CRISPR/Cas9 System 3 Martin L. Nelwan 4 Department of Animal Science – Other 5 Nelwan Institution for Human Resource Development 6 Jl. A. Yani No. 24 7 Palu, Sulawesi Tengah, Indonesia 8 Email: [email protected] 9 Abstract: Antisocial behavior is a behavior disorder inherited according to the inheritance of X-linked 10 chromosome. Mutations in the MAOA gene can cause different behaviors in humans. These can comprise 11 violent behavior or antisocial behavior. Low MAOA (MAOA-L) allele activity can cause antisocial 12 behavior in both healthy and unhealthy people. Antisocial from healthy males can originate from 13 maltreatment during childhood. There are no drugs for the treatment of antisocial behavior permanently 14 at this time. MAOA inhibitor can reverse antisocial behavior in animal models. To cure antisocial 15 behavior in the future, the CRISPR/Cas9 system in combination with iPSCs or ssODN methods for 16 instance can be used. This system has succeeded to correct erroneous segments in the F8 gene and F9 17 gene. Both genes occupy the X chromosome. The MAOA gene also occupies the X chromosome. It seems 18 that CRISPR/Cas9 system may be a beneficial tool to edit erroneous segments in the MAOA gene to treat 19 antisocial behavior. 20 Keywords: advanced therapy, aggressive, antisocial, behavior, MAOA. 21 22 1. Introduction 23 Antisocial behavior is a hereditary disorder inherited through an X-linked recessive inheritance 24 pattern. -

CRASH Syndrome: Clinical Spectrum Vance Lemmon0 Guy Van Campa of Corpus Callosum Hypoplasia, Lieve Vitsa Retardation, Adducted Thumbs, Paul Couckea Patrick J
Review 1608178 Eur J Hum Genet 1995;3:273-284 Erik Fransen3' CRASH Syndrome: Clinical Spectrum Vance Lemmon0 Guy Van Campa of Corpus Callosum Hypoplasia, Lieve Vitsa Retardation, Adducted Thumbs, Paul Couckea Patrick J. Willemsa Spastic Paraparesis and a Department of Medical Genetics, Hydrocephalus Due to Mutations in University of Antwerp, Belgium; One Single Gene, L1 b Case Western Reserve University, Cleveland, Ohio, USA Keywords Abstract X-linked disorder LI is a neuronal cell adhesion molecule with important func Mental retardation tions in the development of the nervous system. The gene Hydrocephalus encoding LI is located near the telomere of the long arm of the MASA syndrome X chromosome in Xq28. We review here the evidence that Adducted thumbs several X-linked mental retardation syndromes including X- Corpus callosum agenesis linked hydrocephalus (HSAS), MASA syndrome, X-linked Spastic paraplegia complicated spastic paraparesis (SPI) and X-linked corpus LI callosum agenesis (ACC) are all due to mutations in the LI CRASH gene. The inter- and intrafamilial variability in families with Mutation analysis an LI mutation is very wide, and patients with HSAS, MASA, SPI and ACC can be present within the same family. There fore, we propose here to refer to this clinical syndrome with the acronym CRASH, for Corpus callosum hypoplasia, Retar dation, Adducted thumbs, Spastic paraplegia and Hydroceph alus. Clinical Aspects for Hydrocephalus due to Stenosis of the Aqueduct of Sylvius. This designation was X-Linked Hydrocephalus based upon the presence of aqueductal steno X-linked hydrocephalus (MIM No. sis in many HSAS patients [1]. However, later 307000) was originally described by Bickers studies reported several HSAS patients with and Adams in 1949. -

Blueprint Genetics Hereditary Leukemia Panel
Hereditary Leukemia Panel Test code: ON0101 Is a 41 gene panel that includes assessment of non-coding variants. Is ideal for patients with a personal history of a syndrome that confers an increased risk of leukemia or patients with a family history of a syndrome that confers an increased risk of leukemia. About Hereditary Leukemia An inherited predisposition to hematological malignancies, namely acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and bone marrow myelodysplastic syndrome (MDS) may be associated with syndromic features or occur as the principal clinical feature. MDSs and AMLs can occur in the context of syndromic bone marrow failure (eg. dyskeratosis congenita, Fanconi anemia). Other hereditary syndromes with an increased risk of leukemia include Li-Fraumeni syndrome (TP53), ataxia telangiectasia (ATM), Bloom syndrome (BLM), neurofibromatosis type 1 (NF1) and less frequently Noonan syndrome (PTPN11, CBL). Some reports have also shown an association of biallelic germline mutations in constitutional mismatch repair-deficiency syndrome genes, MLH1, MSH2, MSH6, and PMS2 with the development of ALL. Isolated hematological malignancies are associated with germline mutations in RUNX1 (familial platelet syndrome with predisposition to acute myelogenous leukemia), CEBPA (familial AML), GATA2 (GATA2-associated syndromes) and DDX41(DDX41 -related myeloid neoplasms). There is a rapidly expanding list of germline mutations associated with increased risks for myeloid malignancies and inherited predisposition to hematologic malignancies may be more common than has been thought. Many different genetic defects associated with the development of leukemia have been described but the common underlying mechanism is a dysfunctional DNA damage response. Recognition of an inherited cause provides a specific molecular diagnosis and helps to guide treatment, understand unique disease features, prognosis and other organ systems that may be involved, and identify others in the family who may be at risk. -

The Mutability and Repair of DNA
B.Sc (Hons) Microbiology (CBCS Structure) C-7: Molecular Biology Unit 2: Replication of DNA The Mutability and Repair of DNA Dr. Rakesh Kumar Gupta Department of Microbiology Ram Lal Anand College New Delhi - 110021 Reference – Molecular Biology of the Gene (6th Edition) by Watson et. al. Pearson education, Inc Principles of Genetics (8th Edition) by D. Peter Snustad, D. Snustad, Eldon Gardner, and Michael J. Simmons, Wiley publications Replication errors and their repair The Nature of Mutations: • Transitions: A kind of the simplest mutation which is pyrimidine-to-pyrimidine and purine-to- purine substitutions such as T to C and A to G • Transversions: The other kind of mutation which are pyrimidine-to-purine and purine-to- pyrimidine substitutions such as T to A or G and ATransitions to C or T. Transversions 8 types 4 types Synonymous substitution Non synonymous substitution • Point mutations: Mutations that alter a single nucleotide Other kinds of mutations (which cause more drastic changes in DNA): • Insertions • Deletions • Gross rearrangements of Thesechromosome mutations might be caused by insertion by transposon or by aberrant action of cellular recombination processes. Mutation • Substitution, deletion, or insertion of a base pair. • Chromosomal deletion, insertion, or rearrangement. Somatic mutations occur in somatic cells and only affect the individual in which the mutation arises. Germ-line mutations alter gametes and passed to the next generation. Mutations are quantified in two ways: 1. Mutation rate = probability of a particular type of mutation per unit time (or generation). 2. Mutation frequency = number of times a particular mutation occurs in a population of cells or individuals. -

Genetic Causes.Pdf
1 September 2015 Genetic causes of childhood apraxia of speech: Case‐based introduction to DNA, inheritance, and clinical management Beate Peter, Ph.D., CCC‐SLP Assistant Professor Dpt. of Speech & Hearing Science Arizona State University Adjunct Assistant Professor AG Dpt. of Communication Sciences & Disorders ATAGCT Saint Louis University T TAGCT Affiliate Assistant Professor Dpt. of Speech & Hearing Sciences University of Washington 1 Disclosure Statement Disclosure Statement Dr. Peter is co‐editor of a textbook on speech development and disorders (B. Peter & A. MacLeod, Eds., 2013), for which she may receive royalty payments. If she shares information about her ongoing research study, this may result in referrals of potential research participants. She has no financial interest or related personal interest of bias in any organization whose products or services are described, reviewed, evaluated or compared in the presentation. 2 Agenda Topic Concepts Why we should care about genetics. Case 1: A sporadic case of CAS who is missing a • Cell, nucleus, chromosomes, genes gene. Introduction to the language of genetics • From genes to proteins • CAS can result when a piece of DNA is deleted or duplicated Case 2: A multigenerational family with CAS • How the FOXP2 gene was discovered and why research in genetics of speech and language disorders is challenging • Pathways from genes to proteins to brain/muscle to speech disorder Case 3: One family's quest for answers • Interprofessional teams, genetic counselors, medical geneticists, research institutes • Early signs of CAS, parent education, early intervention • What about genetic testing? Q&A 3 “Genetic Causes of CAS: Case-Based Introduction to DNA, Inheritance and Clinical Management,” Presented by: Beate Peter, PhD, CCC-SLP, September 29, 2015, Sponsored by: CASANA 2 Why should you care about genetics? 4 If you are a parent of a child with childhood apraxia of speech … 5 When she was in preschool, He doesn’t have any friends. -

Menkes Disease in 5 Siblings
Cu(e) the balancing act: Copper homeostasis explored in 5 siblings Poster with variable clinical course 595 Sonia A Varghese MD MPH MBA and Yael Shiloh-Malawsky MD Objective Methods Case Presentation Discussion v Present a unique variation of v Review literature describing v The graph below depicts the phenotypic spectrum seen in the 5 brothers v ATP7A mutations produce a clinical spectrum phenotype and course in siblings copper transport disorders v Mom is a known carrier of ATP7A mutation v Siblings 1, 2, and 5 follow a more classic with Menkes Disease (MD) v Apply findings to our case of five v There is limited information on the siblings who were not seen at UNC Menke’s course, while siblings 3 and 4 exhibit affected siblings v The 6th sibling is the youngest and is a healthy female infant (not included here) a phenotypic variation Sibling: v This variation is suggestive of a milder form of Background Treatment birth Presentation Exam Diagnostic testing Outcomes Copper Histidine Menkes such as occipital horn syndrome with v Mutations in ATP7A: copper deficiency order D: 16mos residual copper transport function (Menkes disease) O/AD: 1 Infancy/12mos N/A N/A No Brain v Siblings 3 and 4 had improvement with copper v Mutations in ATP7B: copper overload FTT, Seizures, DD hemorrhage supplementation, however declined when off (Wilson disease) O/AD: Infancy/NA D: 13mos supplementation -suggesting residual ATP7A v The amount of residual functioning 2 N/A N/A No FTT, FTT copper transport function copper transport influences disease Meningitis -

Exostoses, Enchondromatosis and Metachondromatosis; Diagnosis and Management
Acta Orthop. Belg., 2016, 82, 102-105 ORIGINAL STUDY Exostoses, enchondromatosis and metachondromatosis; diagnosis and management John MCFARLANE, Tim KNIGHT, Anubha SINHA, Trevor COLE, Nigel KIELY, Rob FREEMAN From the Department of Orthopaedics, Robert Jones Agnes Hunt Hospital, Oswestry, UK We describe a 5 years old girl who presented to the region of long bones and are composed of a carti- multidisciplinary skeletal dysplasia clinic following lage lump outside the bone which may be peduncu- excision of two bony lumps from her fingers. Based on lated or sessile, the knee is the most common clinical examination, radiolographs and histological site (1,10). An isolated exostosis is a common inci- results an initial diagnosis of hereditary multiple dental finding rarely requiring treatment. Disorders exostosis (HME) was made. Four years later she developed further lumps which had the radiological associated with exostoses include HME, Langer- appearance of enchondromas. The appearance of Giedion syndrome, Gardner syndrome and meta- both exostoses and enchondromas suggested a possi- chondromatosis. ble diagnosis of metachondromatosis. Genetic testing Enchondroma are the second most common be- revealed a splice site mutation at the end of exon 11 on nign bone tumour characterised by the formation of the PTPN11 gene, confirming the diagnosis of meta- hyaline cartilage in the medulla of a bone. It occurs chondromatosis. While both single or multiple exosto- most frequently in the hand (60%) and then the feet. ses and enchondromas occur relatively commonly on The typical radiological features are of a well- their own, the appearance of multiple exostoses and defined lucent defect with endosteal scalloping and enchondromas together is rare and should raise the differential diagnosis of metachondromatosis. -

Hereditary Spastic Paraparesis: a Review of New Developments
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.69.2.150 on 1 August 2000. Downloaded from 150 J Neurol Neurosurg Psychiatry 2000;69:150–160 REVIEW Hereditary spastic paraparesis: a review of new developments CJ McDermott, K White, K Bushby, PJ Shaw Hereditary spastic paraparesis (HSP) or the reditary spastic paraparesis will no doubt Strümpell-Lorrain syndrome is the name given provide a more useful and relevant classifi- to a heterogeneous group of inherited disorders cation. in which the main clinical feature is progressive lower limb spasticity. Before the advent of Epidemiology molecular genetic studies into these disorders, The prevalence of HSP varies in diVerent several classifications had been proposed, studies. Such variation is probably due to a based on the mode of inheritance, the age of combination of diVering diagnostic criteria, onset of symptoms, and the presence or other- variable epidemiological methodology, and wise of additional clinical features. Families geographical factors. Some studies in which with autosomal dominant, autosomal recessive, similar criteria and methods were employed and X-linked inheritance have been described. found the prevalance of HSP/100 000 to be 2.7 in Molise Italy, 4.3 in Valle d’Aosta Italy, and 10–12 Historical aspects 2.0 in Portugal. These studies employed the In 1880 Strümpell published what is consid- diagnostic criteria suggested by Harding and ered to be the first clear description of HSP.He utilised all health institutions and various reported a family in which two brothers were health care professionals in ascertaining cases aVected by spastic paraplegia. The father was from the specific region. -
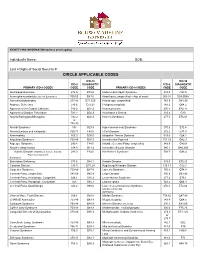
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

My Beloved Neutrophil Dr Boxer 2014 Neutropenia Family Conference
The Beloved Neutrophil: Its Function in Health and Disease Stem Cell Multipotent Progenitor Myeloid Lymphoid CMP IL-3, SCF, GM-CSF CLP Committed Progenitor MEP GMP GM-CSF, IL-3, SCF EPO TPO G-CSF M-CSF IL-5 IL-3 SCF RBC Platelet Neutrophil Monocyte/ Basophil B-cells Macrophage Eosinophil T-Cells Mast cell NK cells Mature Cell Dendritic cells PRODUCTION AND KINETICS OF NEUTROPHILS CELLS % CELLS TIME Bone Marrow: Myeloblast 1 7 - 9 Mitotic Promyelocyte 4 Days Myelocyte 16 Maturation/ Metamyelocyte 22 3 – 7 Storage Band 30 Days Seg 21 Vascular: Peripheral Blood Seg 2 6 – 12 hours 3 Marginating Pool Apoptosis and ? Tissue clearance by 0 – 3 macrophages days PHAGOCYTOSIS 1. Mobilization 2. Chemotaxis 3. Recognition (Opsonization) 4. Ingestion 5. Degranulation 6. Peroxidation 7. Killing and Digestion 8. Net formation Adhesion: β 2 Integrins ▪ Heterodimer of a and b chain ▪ Tight adhesion, migration, ingestion, co- stimulation of other PMN responses LFA-1 Mac-1 (CR3) p150,95 a2b2 a CD11a CD11b CD11c CD11d b CD18 CD18 CD18 CD18 Cells All PMN, Dendritic Mac, mono, leukocytes mono/mac, PMN, T cell LGL Ligands ICAMs ICAM-1 C3bi, ICAM-3, C3bi other other Fibrinogen other GRANULOCYTE CHEMOATTRACTANTS Chemoattractants Source Activators Lipids PAF Neutrophils C5a, LPS, FMLP Endothelium LTB4 Neutrophils FMLP, C5a, LPS Chemokines (a) IL-8 Monocytes, endothelium LPS, IL-1, TNF, IL-3 other cells Gro a, b, g Monocytes, endothelium IL-1, TNF other cells NAP-2 Activated platelets Platelet activation Others FMLP Bacteria C5a Activation of complement Other Important Receptors on PMNs ñ Pattern recognition receptors – Detect microbes - Toll receptor family - Mannose receptor - bGlucan receptor – fungal cell walls ñ Cytokine receptors – enhance PMN function - G-CSF, GM-CSF - TNF Receptor ñ Opsonin receptors – trigger phagocytosis - FcgRI, II, III - Complement receptors – ñ Mac1/CR3 (CD11b/CD18) – C3bi ñ CR-1 – C3b, C4b, C3bi, C1q, Mannose binding protein From JG Hirsch, J Exp Med 116:827, 1962, with permission. -

MASA Syndrome in Twin Brothers: Case Report of Sixteen-Year Clinical Follow Up
Paediatr Croat. 2014;58:286-90 PRIKAZ BOLESNIKA / CASE REPORT www.paedcro.com http://dx.doi.org/10.13112/PC.2014.50 MASA syndrome in twin brothers: case report of sixteen-year clinical follow up Matilda Kovač Šižgorić1, Zlatko Sabol1, Filip Sabol2, Tonći Grmoja3, Svjetlana Bela Klancir1, Zdravka Gjergja1, Ljiljana Kipke Sabol1 MASA syndrome (OMIM 303350) is a rare X-linked recessive neurologic disorder, also called CRASH syndrome, spastic paraplegia 1 and Gareis-Mason syndrome. The acronym MASA describes four major signs: Mental retardation, Aphasia, Shuffl ing gait and Adducted thumbs. A more suitable name for this syndrome is L1 syndrome because the disorder has been associated with mutations in the neuronal cell adhesion molecule L1 (L1CAM) gene. The syndrome has severe symptoms in males, while females are carriers because only one X chromosome is aff ected. The aim of this report is to show similarities and diff erences in clinical manifestations between twins with the L1CAM gene mutation and to emphasize the importance of genetic counseling. Our patients were dizygotic twins born prematurely at 35 weeks of gestation. Pregnancy was complicated with early bleeding and gestational diabetes. Immediately after birth, hypertonia of lower extremities was observed in both twins. Sixteen-year clinical follow up showed spastic paraparetic form with shuffl ing gait, clumsiness, delayed speech development, lower intellectual functioning at the level of mild to moderate mental retarda- tion, primary nocturnal enuresis, behavioral and sleep disorder (more pronounced in the second twin). Magnetic resonance imaging of the brain showed complete agenesis of the corpus callosum, complete lack of the anterior commissure, and internal hydrocephalus.