Somatic Mutations in the Human Brain: Implications for Psychiatric Research
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Paternal Factors and Schizophrenia Risk: De Novo Mutations and Imprinting
Paternal Factors and Schizophrenia Risk: De Novo Mutations and Imprinting by Dolores Malaspina Downloaded from https://academic.oup.com/schizophreniabulletin/article/27/3/379/1835092 by guest on 23 September 2021 Abstract (Impagnatiello et al. 1998; Kao et al. 1998), but there is no consensus that any particular gene plays a meaning- There is a strong genetic component for schizophrenia ful role in the etiology of schizophrenia (Hyman 2000). risk, but it is unclear how the illness is maintained in Some of the obstacles in genetic research in schiz- the population given the significantly reduced fertility ophrenia are those of any complex disorder, and include of those with the disorder. One possibility is that new incomplete penetrance, polygenic interaction (epista- mutations occur in schizophrenia vulnerability genes. sis), diagnostic instability, and variable expressivity. If so, then those with schizophrenia may have older Schizophrenia also does not show a clear Mendelian fathers, because advancing paternal age is the major inheritance pattern, although segregation analyses have source of new mutations in humans. This review variably supported dominant, recessive, additive, sex- describes several neurodevelopmental disorders that linked, and oligogenic inheritance (Book 1953; Slater have been associated with de novo mutations in the 1958; Garrone 1962; Elston and Campbell 1970; Slater paternal germ line and reviews data linking increased and Cowie 1971; Karlsson 1972; Stewart et al. 1980; schizophrenia risk with older fathers. Several genetic Risch 1990a, 1990fc; reviewed by Kendler and Diehl mechanisms that could explain this association are 1993). Furthermore, both nonallelic (Kaufmann et al. proposed, including paternal germ line mutations, 1998) and etiologic heterogeneity (Malaspina et al. -

Bhagwan Moorjani, MD, FAAP, FAAN • Requires Knowledge of Normal CNS Developmental (I.E
1/16/2012 Neuroimaging in Childhood • Neuroimaging issues are distinct from Pediatric Neuroimaging in adults Neurometabolic-degenerative disorder • Sedation/anesthesia and Epilepsy • Motion artifacts Bhagwan Moorjani, MD, FAAP, FAAN • Requires knowledge of normal CNS developmental (i.e. myelin maturation) • Contrast media • Parental anxiety Diagnostic Approach Neuroimaging in Epilepsy • Age of onset • Peak incidence in childhood • Static vs Progressive • Occurs as a co-morbid condition in many – Look for treatable causes pediatric disorders (birth injury, – Do not overlook abuse, Manchausen if all is negative dysmorphism, chromosomal anomalies, • Phenotype presence (syndromic, HC, NCS, developmental delays/regression) systemic involvement) • Predominant symptom (epilepsy, DD, • Many neurologic disorders in children weakness/motor, psychomotor regression, have the same chief complaint cognitive/dementia) 1 1/16/2012 Congenital Malformation • Characterized by their anatomic features • Broad categories: based on embryogenesis – Stage 1: Dorsal Induction: Formation and closure of the neural tube. (Weeks 3-4) – Stage 2: Ventral Induction: Formation of the brain segments and face. (Weeks 5-10) – Stage 3: Migration and Histogenesis: (Months 2-5) – Stage 4: Myelination: (5-15 months; matures by 3 years) Dandy Walker Malformation Dandy walker • Criteria: – high position of tentorium – dysgenesis/agenesis of vermis – cystic dilatation of fourth ventricle • commonly associated features: – hypoplasia of cerebellum – scalloping of inner table of occipital bone • associated abnormalities: – hydrocephalus 75% – dysgenesis of corpus callosum 25% – heterotropia 10% 2 1/16/2012 Etiology of Epilepsy: Developmental and Genetic Classification of Gray Matter Heterotropia Cortical Dysplasia 1. Secondary to abnormal neuronal and • displaced masses of nerve cells • Subependymal glial proliferation/apoptosis (gray matter) heterotropia (most • most common: small nest common) 2. -

Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin
cells Review Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin Agnieszka Bochy ´nska,Juliane Lüscher-Firzlaff and Bernhard Lüscher * ID Institute of Biochemistry and Molecular Biology, Medical School, RWTH Aachen University, Pauwelsstrasse 30, 52057 Aachen, Germany; [email protected] (A.B.); jluescher-fi[email protected] (J.L.-F.) * Correspondence: [email protected]; Tel.: +49-241-8088850; Fax: +49-241-8082427 Received: 18 January 2018; Accepted: 27 February 2018; Published: 2 March 2018 Abstract: Regulation of gene expression is achieved by sequence-specific transcriptional regulators, which convey the information that is contained in the sequence of DNA into RNA polymerase activity. This is achieved by the recruitment of transcriptional co-factors. One of the consequences of co-factor recruitment is the control of specific properties of nucleosomes, the basic units of chromatin, and their protein components, the core histones. The main principles are to regulate the position and the characteristics of nucleosomes. The latter includes modulating the composition of core histones and their variants that are integrated into nucleosomes, and the post-translational modification of these histones referred to as histone marks. One of these marks is the methylation of lysine 4 of the core histone H3 (H3K4). While mono-methylation of H3K4 (H3K4me1) is located preferentially at active enhancers, tri-methylation (H3K4me3) is a mark found at open and potentially active promoters. Thus, H3K4 methylation is typically associated with gene transcription. The class 2 lysine methyltransferases (KMTs) are the main enzymes that methylate H3K4. KMT2 enzymes function in complexes that contain a necessary core complex composed of WDR5, RBBP5, ASH2L, and DPY30, the so-called WRAD complex. -

Inherited Neuropathies
407 Inherited Neuropathies Vera Fridman, MD1 M. M. Reilly, MD, FRCP, FRCPI2 1 Department of Neurology, Neuromuscular Diagnostic Center, Address for correspondence Vera Fridman, MD, Neuromuscular Massachusetts General Hospital, Boston, Massachusetts Diagnostic Center, Massachusetts General Hospital, Boston, 2 MRC Centre for Neuromuscular Diseases, UCL Institute of Neurology Massachusetts, 165 Cambridge St. Boston, MA 02114 and The National Hospital for Neurology and Neurosurgery, Queen (e-mail: [email protected]). Square, London, United Kingdom Semin Neurol 2015;35:407–423. Abstract Hereditary neuropathies (HNs) are among the most common inherited neurologic Keywords disorders and are diverse both clinically and genetically. Recent genetic advances have ► hereditary contributed to a rapid expansion of identifiable causes of HN and have broadened the neuropathy phenotypic spectrum associated with many of the causative mutations. The underlying ► Charcot-Marie-Tooth molecular pathways of disease have also been better delineated, leading to the promise disease for potential treatments. This chapter reviews the clinical and biological aspects of the ► hereditary sensory common causes of HN and addresses the challenges of approaching the diagnostic and motor workup of these conditions in a rapidly evolving genetic landscape. neuropathy ► hereditary sensory and autonomic neuropathy Hereditary neuropathies (HN) are among the most common Select forms of HN also involve cranial nerves and respiratory inherited neurologic diseases, with a prevalence of 1 in 2,500 function. Nevertheless, in the majority of patients with HN individuals.1,2 They encompass a clinically heterogeneous set there is no shortening of life expectancy. of disorders and vary greatly in severity, spanning a spectrum Historically, hereditary neuropathies have been classified from mildly symptomatic forms to those resulting in severe based on the primary site of nerve pathology (myelin vs. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Mutation in Genes FBN1, AKT1, and LMNA: Marfan Syndrome, Proteus Syndrome, and Progeria Share Common Systemic Involvement
Review Mutation in Genes FBN1, AKT1, and LMNA: Marfan Syndrome, Proteus Syndrome, and Progeria Share Common Systemic Involvement Tonmoy Biswas.1 Abstract Genetic mutations are becoming more deleterious day by day. Mutations of Genes named FBN1, AKT1, LMNA result specific protein malfunction that in turn commonly cause Marfan syndrome, Proteus syndrome, and Progeria, respectively. Articles about these conditions have been reviewed in PubMed and Google scholar with a view to finding relevant clinical features. Precise keywords have been used in search for systemic involvement of FBN1, AKT1, and LMNA gene mutations. It has been found that Marfan syndrome, Proteus syndrome, and Progeria commonly affected musculo-skeletal system, cardiovascular system, eye, and nervous system. Not only all of them shared identical systemic involvement, but also caused several very specific anomalies in various parts of the body. In spite of having some individual signs and symptoms, the mutual manifestations were worth mentio- ning. Moreover, all the features of the mutations of all three responsible genes had been co-related and systemically mentioned in this review. There can be some mutual properties of the genes FBN1, AKT1, and LMNA or in their corresponding proteins that result in the same presentations. This study may progress vision of knowledge regarding risk factors, patho-physiology, and management of these conditions, and relation to other mutations. Keywords: Genetic mutation; Marfan syndrome; Proteus syndrome; Progeria; Gene FBN1; Gene AKT1; Gene LMNA; Musculo-skeletal system; Cardiovascular system; Eye; Nervous system (Source: MeSH, NLM). Introduction Records in human mutation databases are increasing day by 5 About the author: Tonmoy The haploid human genome consists of 3 billion nucleotides day. -
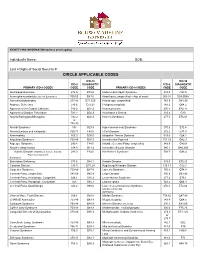
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

2018 Etiologies by Frequencies
2018 Etiologies in Order of Frequency by Category Hereditary Syndromes and Disorders Count CHARGE Syndrome 958 Down syndrome (Trisomy 21 syndrome) 308 Usher I syndrome 252 Stickler syndrome 130 Dandy Walker syndrome 119 Cornelia de Lange 102 Goldenhar syndrome 98 Usher II syndrome 83 Wolf-Hirschhorn syndrome (Trisomy 4p) 68 Trisomy 13 (Trisomy 13-15, Patau syndrome) 60 Pierre-Robin syndrome 57 Moebius syndrome 55 Trisomy 18 (Edwards syndrome) 52 Norrie disease 38 Leber congenital amaurosis 35 Chromosome 18, Ring 18 31 Aicardi syndrome 29 Alstrom syndrome 27 Pfieffer syndrome 27 Treacher Collins syndrome 27 Waardenburg syndrome 27 Marshall syndrome 25 Refsum syndrome 21 Cri du chat syndrome (Chromosome 5p- synd) 16 Bardet-Biedl syndrome (Laurence Moon-Biedl) 15 Hurler syndrome (MPS I-H) 15 Crouzon syndrome (Craniofacial Dysotosis) 13 NF1 - Neurofibromatosis (von Recklinghausen dis) 13 Kniest Dysplasia 12 Turner syndrome 11 Usher III syndrome 10 Cockayne syndrome 9 Apert syndrome/Acrocephalosyndactyly, Type 1 8 Leigh Disease 8 Alport syndrome 6 Monosomy 10p 6 NF2 - Bilateral Acoustic Neurofibromatosis 6 Batten disease 5 Kearns-Sayre syndrome 5 Klippel-Feil sequence 5 Hereditary Syndromes and Disorders Count Prader-Willi 5 Sturge-Weber syndrome 5 Marfan syndrome 3 Hand-Schuller-Christian (Histiocytosis X) 2 Hunter Syndrome (MPS II) 2 Maroteaux-Lamy syndrome (MPS VI) 2 Morquio syndrome (MPS IV-B) 2 Optico-Cochleo-Dentate Degeneration 2 Smith-Lemli-Opitz (SLO) syndrome 2 Wildervanck syndrome 2 Herpes-Zoster (or Hunt) 1 Vogt-Koyanagi-Harada -

Clinical Utility of Recently Identified Diagnostic, Prognostic, And
Modern Pathology (2017) 30, 1338–1366 1338 © 2017 USCAP, Inc All rights reserved 0893-3952/17 $32.00 Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms Arantza Onaindia1, L Jeffrey Medeiros2 and Keyur P Patel2 1Instituto de Investigacion Marques de Valdecilla (IDIVAL)/Hospital Universitario Marques de Valdecilla, Santander, Spain and 2Department of Hematopathology, MD Anderson Cancer Center, Houston, TX, USA Genomic profiling studies have provided new insights into the pathogenesis of mature B-cell neoplasms and have identified markers with prognostic impact. Recurrent mutations in tumor-suppressor genes (TP53, BIRC3, ATM), and common signaling pathways, such as the B-cell receptor (CD79A, CD79B, CARD11, TCF3, ID3), Toll- like receptor (MYD88), NOTCH (NOTCH1/2), nuclear factor-κB, and mitogen activated kinase signaling, have been identified in B-cell neoplasms. Chronic lymphocytic leukemia/small lymphocytic lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, Burkitt lymphoma, Waldenström macroglobulinemia, hairy cell leukemia, and marginal zone lymphomas of splenic, nodal, and extranodal types represent examples of B-cell neoplasms in which novel molecular biomarkers have been discovered in recent years. In addition, ongoing retrospective correlative and prospective outcome studies have resulted in an enhanced understanding of the clinical utility of novel biomarkers. This progress is reflected in the 2016 update of the World Health Organization classification of lymphoid neoplasms, which lists as many as 41 mature B-cell neoplasms (including provisional categories). Consequently, molecular genetic studies are increasingly being applied for the clinical workup of many of these neoplasms. In this review, we focus on the diagnostic, prognostic, and/or therapeutic utility of molecular biomarkers in mature B-cell neoplasms. -

Megalencephaly and Macrocephaly
277 Megalencephaly and Macrocephaly KellenD.Winden,MD,PhD1 Christopher J. Yuskaitis, MD, PhD1 Annapurna Poduri, MD, MPH2 1 Department of Neurology, Boston Children’s Hospital, Boston, Address for correspondence Annapurna Poduri, Epilepsy Genetics Massachusetts Program, Division of Epilepsy and Clinical Electrophysiology, 2 Epilepsy Genetics Program, Division of Epilepsy and Clinical Department of Neurology, Fegan 9, Boston Children’s Hospital, 300 Electrophysiology, Department of Neurology, Boston Children’s Longwood Avenue, Boston, MA 02115 Hospital, Boston, Massachusetts (e-mail: [email protected]). Semin Neurol 2015;35:277–287. Abstract Megalencephaly is a developmental disorder characterized by brain overgrowth secondary to increased size and/or numbers of neurons and glia. These disorders can be divided into metabolic and developmental categories based on their molecular etiologies. Metabolic megalencephalies are mostly caused by genetic defects in cellular metabolism, whereas developmental megalencephalies have recently been shown to be caused by alterations in signaling pathways that regulate neuronal replication, growth, and migration. These disorders often lead to epilepsy, developmental disabilities, and Keywords behavioral problems; specific disorders have associations with overgrowth or abnor- ► megalencephaly malities in other tissues. The molecular underpinnings of many of these disorders are ► hemimegalencephaly now understood, providing insight into how dysregulation of critical pathways leads to ► -

Aberrant Activity of Histone–Lysine N-Methyltransferase 2 (KMT2) Complexes in Oncogenesis
International Journal of Molecular Sciences Review Aberrant Activity of Histone–Lysine N-Methyltransferase 2 (KMT2) Complexes in Oncogenesis Elzbieta Poreba 1,* , Krzysztof Lesniewicz 2 and Julia Durzynska 1,* 1 Institute of Experimental Biology, Faculty of Biology, Adam Mickiewicz University, ul. Uniwersytetu Pozna´nskiego6, 61-614 Pozna´n,Poland 2 Department of Molecular and Cellular Biology, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, ul. Uniwersytetu Pozna´nskiego6, 61-614 Pozna´n,Poland; [email protected] * Correspondence: [email protected] (E.P.); [email protected] (J.D.); Tel.: +48-61-829-5857 (E.P.) Received: 19 November 2020; Accepted: 6 December 2020; Published: 8 December 2020 Abstract: KMT2 (histone-lysine N-methyltransferase subclass 2) complexes methylate lysine 4 on the histone H3 tail at gene promoters and gene enhancers and, thus, control the process of gene transcription. These complexes not only play an essential role in normal development but have also been described as involved in the aberrant growth of tissues. KMT2 mutations resulting from the rearrangements of the KMT2A (MLL1) gene at 11q23 are associated with pediatric mixed-lineage leukemias, and recent studies demonstrate that KMT2 genes are frequently mutated in many types of human cancers. Moreover, other components of the KMT2 complexes have been reported to contribute to oncogenesis. This review summarizes the recent advances in our knowledge of the role of KMT2 complexes in cell transformation. In addition, it discusses the therapeutic targeting of different components of the KMT2 complexes. Keywords: histone–lysine N-methyltransferase 2; COMPASS; COMPASS-like; H3K4 methylation; oncogenesis; cancer; epigenetics; chromatin 1.