3. Evolution Makes Sense of Homologies Richard Owen (1848
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
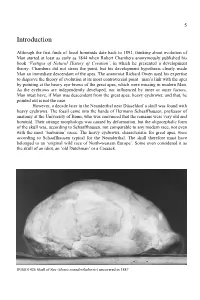
Introduction
5 Introduction Although the first finds of fossil hominids date back to 1891, thinking about evolution of Man started at least as early as 1844 when Robert Chambers anonymously published his book ‘Vestiges of Natural History of Creation’, in which he presented a development theory. Chambers did not stress the point, but his development hypothesis clearly made Man an immediate descendant of the apes. The anatomist Richard Owen used his expertise to disprove the theory of evolution at its most controversial point –man’s link with the apes by pointing at the heavy eye-brows of the great apes, which were missing in modern Man. As the eyebrows are independently developed, nor influenced by inner or outer factors, Man must have, if Man was descendent from the great apes, heavy eyebrows; and that, he pointed out is not the case. However, a decade later in the Neanderthal near Düsseldorf a skull was found with heavy eyebrows. The fossil came into the hands of Hermann Schaaffhausen, professor of anatomy at the University of Bonn, who was convinced that the remains were very old and hominid. Their strange morphology was caused by deformation, but the oligocephalic form of the skull was, according to Schaaffhausen, not comparable to any modern race, not even with the most ‘barbarian’ races. The heavy eyebrows, characteristic for great apes, were according to Schaaffhausen typical for the Neanderthal. The skull therefore must have belonged to an ‘original wild race of North-western Europe’. Some even considered it as the skull of an idiot, an ‘old Dutchman’ or a Cossack. -

The Correspondence of Julius Haast and Joseph Dalton Hooker, 1861-1886
The Correspondence of Julius Haast and Joseph Dalton Hooker, 1861-1886 Sascha Nolden, Simon Nathan & Esme Mildenhall Geoscience Society of New Zealand miscellaneous publication 133H November 2013 Published by the Geoscience Society of New Zealand Inc, 2013 Information on the Society and its publications is given at www.gsnz.org.nz © Copyright Simon Nathan & Sascha Nolden, 2013 Geoscience Society of New Zealand miscellaneous publication 133H ISBN 978-1-877480-29-4 ISSN 2230-4495 (Online) ISSN 2230-4487 (Print) We gratefully acknowledge financial assistance from the Brian Mason Scientific and Technical Trust which has provided financial support for this project. This document is available as a PDF file that can be downloaded from the Geoscience Society website at: http://www.gsnz.org.nz/information/misc-series-i-49.html Bibliographic Reference Nolden, S.; Nathan, S.; Mildenhall, E. 2013: The Correspondence of Julius Haast and Joseph Dalton Hooker, 1861-1886. Geoscience Society of New Zealand miscellaneous publication 133H. 219 pages. The Correspondence of Julius Haast and Joseph Dalton Hooker, 1861-1886 CONTENTS Introduction 3 The Sumner Cave controversy Sources of the Haast-Hooker correspondence Transcription and presentation of the letters Acknowledgements References Calendar of Letters 8 Transcriptions of the Haast-Hooker letters 12 Appendix 1: Undated letter (fragment), ca 1867 208 Appendix 2: Obituary for Sir Julius von Haast 209 Appendix 3: Biographical register of names mentioned in the correspondence 213 Figures Figure 1: Photographs -

Chapter 10, the Mistaken Extinction, by Lowell Dingus and Timothy Rowe, New York, W
Chapter 10, The Mistaken Extinction, by Lowell Dingus and Timothy Rowe, New York, W. H. Freeman, 1998. CHAPTER 10 Dinosaurs Challenge Evolution Enter Sir Richard Owen More than 150 years ago, the great British naturalist Richard Owen (fig. 10.01) ignited the controversy that Deinonychus would eventually inflame. The word "dinosaur" was first uttered by Owen in a lecture delivered at Plymouth, England in July of 1841. He had coined the name in a report on giant fossil reptiles that were discovered in England earlier in the century. The root, Deinos, is usually translated as "terrible" but in his report, published in 1842, Owen chose the words "fearfully great"1. To Owen, dinosaurs were the fearfully great saurian reptiles, known only from fossil skeletons of huge extinct animals, unlike anything alive today. Fig. 10.01 Richard Owen as, A) a young man at about the time he named Dinosauria, B) in middle age, near the time he described Archaeopteryx, and C) in old age. Dinosaur bones were discovered long before Owen first spoke their name, but no one understood what they represented. The first scientific report on a dinosaur bone belonging was printed in 1677 by Rev. Robert Plot in his work, The Natural History of Oxfordshire. This broken end of a thigh bone, came to Plot's attention during his research. It was nearly 60 cm in circumference--greater than the same bone in an elephant (fig.10.02). We now suspect that it belonged to Megalosaurus bucklandii, a carnivorous dinosaur now known from Oxfordshire. But Plot concluded that it "must have been a real Bone, now petrified" and that it resembled "exactly the figure of the 1 Chapter 10, The Mistaken Extinction, by Lowell Dingus and Timothy Rowe, New York, W. -

Designing the Dinosaur: Richard Owen's Response to Robert Edmond Grant Author(S): Adrian J
Designing the Dinosaur: Richard Owen's Response to Robert Edmond Grant Author(s): Adrian J. Desmond Source: Isis, Vol. 70, No. 2 (Jun., 1979), pp. 224-234 Published by: The University of Chicago Press on behalf of The History of Science Society Stable URL: http://www.jstor.org/stable/230789 . Accessed: 16/10/2013 13:00 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp . JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. The University of Chicago Press and The History of Science Society are collaborating with JSTOR to digitize, preserve and extend access to Isis. http://www.jstor.org This content downloaded from 150.135.115.18 on Wed, 16 Oct 2013 13:00:27 PM All use subject to JSTOR Terms and Conditions Designing the Dinosaur: Richard Owen's Response to Robert Edmond Grant By Adrian J. Desmond* I N THEIR PAPER on "The Earliest Discoveries of Dinosaurs" Justin Delair and William Sarjeant permit Richard Owen to step in at the last moment and cap two decades of frenzied fossil collecting with the word "dinosaur."' This approach, I believe, denies Owen's real achievement while leaving a less than fair impression of the creative aspect of science. -

Marsupials As Ancestors Or Sister Taxa?
Archives of natural history 39.2 (2012): 217–233 Edinburgh University Press DOI: 10.3366/anh.2012.0091 # The Society for the History of Natural History www.eupjournals.com/anh Darwin’s two competing phylogenetic trees: marsupials as ancestors or sister taxa? J. DAVID ARCHIBALD Department of Biology, San Diego State University, San Diego, CA 92182–4614, USA (e-mail: [email protected]). ABSTRACT: Studies of the origin and diversification of major groups of plants and animals are contentious topics in current evolutionary biology. This includes the study of the timing and relationships of the two major clades of extant mammals – marsupials and placentals. Molecular studies concerned with marsupial and placental origin and diversification can be at odds with the fossil record. Such studies are, however, not a recent phenomenon. Over 150 years ago Charles Darwin weighed two alternative views on the origin of marsupials and placentals. Less than a year after the publication of On the origin of species, Darwin outlined these in a letter to Charles Lyell dated 23 September 1860. The letter concluded with two competing phylogenetic diagrams. One showed marsupials as ancestral to both living marsupials and placentals, whereas the other showed a non-marsupial, non-placental as being ancestral to both living marsupials and placentals. These two diagrams are published here for the first time. These are the only such competing phylogenetic diagrams that Darwin is known to have produced. In addition to examining the question of mammalian origins in this letter and in other manuscript notes discussed here, Darwin confronted the broader issue as to whether major groups of animals had a single origin (monophyly) or were the result of “continuous creation” as advocated for some groups by Richard Owen. -

BIOL 1406 Darwin's Dangerous Idea
BIOL 1406 Darwin’s Dangerous Idea - Video Exam I Essay Question: (Matching Format) Describe the history of the scientific theory, biological evolution by means of natural selection: and focus on the life of Charles Darwin as portrayed in the PBS production, Darwin’s Dangerous Idea. Be sure to describe the roles of the following: "Raz", Robert FitzRoy, Emma Darwin, Annie Darwin, Richard Owen, Charles Lyell, Thomas Malthus, Samuel Wilberforce, and Thomas Huxley.) 1.Describe Captain Fitzroy’s perspective when it comes to “free-thinking” 2. What does Fitzroy allow Darwin to borrow? 3. Who was “Raz”? 4. Who was Richard Owen? 5. What was Owen’s view on “free-thinking” with regard to human ancestory? 6. What was Owen so afraid of? 7. Who was Emma (Wedgewood) Darwin? How did she influence Charles Darwin with regard to his scientific inquiry ? 8. What type of disease do we now speculate that Darwin may have suffered from? How did he get the disease? 9. Who was Annie Darwin? 10. When Annie left, what affect did this have on Darwin? 11. Who was Charles Lyell? What role did he play in influencing Darwin? 12. Who was Thomas Malthus? What did he do to influence Darwin? 13. What did Richard Owen do that was scientifically unethical? Why did he do this? 14. Who was Samuel Wilberforce? 15. Who was Thomas Henry Huxley? What did he do to influence Darwin? 16. Who was Alfred Russel Wallace? What did he do to influence Darwin? 17. What motivated Darwin to study so many different organisms; i.e. -

Styles of Reasoning in Early to Mid-Victorian Life Research: Analysis:Synthesis and Palaetiology
Journal of the History of Biology (2006) Ó Springer 2006 DOI 10.1007/s10739-006-0006-4 Styles of Reasoning in Early to Mid-Victorian Life Research: Analysis:Synthesis and Palaetiology JAMES ELWICK Science and Technology Studies Faculties of Arts and Science and Engineering York University 4700 Keele St. M3J 1P3 Toronto, ON Canada E-mail: [email protected] Abstract. To better understand the work of pre-Darwinian British life researchers in their own right, this paper discusses two different styles of reasoning. On the one hand there was analysis:synthesis, where an organism was disintegrated into its constituent parts and then reintegrated into a whole; on the other hand there was palaetiology, the historicist depiction of the progressive specialization of an organism. This paper shows how each style allowed for development, but showed it as moving in opposite directions. In analysis:synthesis, development proceeded centripetally, through the fusion of parts. Meanwhile in palaetiology, development moved centrifugally, through the ramifying specialization of an initially simple substance. I first examine a com- munity of analytically oriented British life researchers, exemplified by Richard Owen, and certain technical questions they considered important. These involved the neu- rosciences, embryology, and reproduction and regeneration. The paper then looks at a new generation of British palaetiologists, exemplified by W.B. Carpenter and T.H. Huxley, who succeeded at portraying analysts’ questions as irrelevant. The link between styles of reasoning and physical sites is also explored. Analysts favored museums, which facilitated the examination and display of unchanging marine organisms while providing a power base for analysts. I suggest that palaetiologists were helped by vivaria, which included marine aquaria and Wardian cases. -

Miranda, 5 | 2011 Jim Endersby, Imperial Nature: Joseph Hooker and the Practices of Victorian S
Miranda Revue pluridisciplinaire du monde anglophone / Multidisciplinary peer-reviewed journal on the English- speaking world 5 | 2011 South and Race / Staging Mobility in the United States Jim Endersby, Imperial Nature: Joseph Hooker and the Practices of Victorian Science Laurence Talairach-Vielmas Édition électronique URL : http://journals.openedition.org/miranda/2550 ISSN : 2108-6559 Éditeur Université Toulouse - Jean Jaurès Référence électronique Laurence Talairach-Vielmas, « Jim Endersby, Imperial Nature: Joseph Hooker and the Practices of Victorian Science », Miranda [En ligne], 5 | 2011, mis en ligne le 29 novembre 2011, consulté le 25 octobre 2018. URL : http://journals.openedition.org/miranda/2550 Ce document a été généré automatiquement le 25 octobre 2018. Miranda is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. Jim Endersby, Imperial Nature: Joseph Hooker and the Practices of Victorian S... 1 Jim Endersby, Imperial Nature: Joseph Hooker and the Practices of Victorian Science Laurence Talairach-Vielmas RÉFÉRENCE Jim Endersby, Imperial Nature : Joseph Hooker and the Practices of Victorian Science (Chicago and London : The University of Chicago Press, [2008] 2010), 429 p, ISBN 978–0–226–20791– 9 1 L’ouvrage de Jim Endersby, Imperial Nature : Joseph Hooker and the Practices of Victorian Science, ne se veut pas une biographie du naturaliste Joseph Dalton Hooker (1817–1911). Bien au contraire. Hooker fut un scientifique véritablement victorien, dont la carrière retrace l’évolution du statut de scientifique, les enjeux et tensions au cœur de la profession et les liens entre scientifiques et naturalistes amateurs. L’étude de Endersby se concentre sur les années charnières de la carrière de Hooker, avant son accession à la tête de Kew Gardens, à un moment où le statut du scientifique est en pleine évolution. -

Sir Richard Owen
Dinosaurs today Year 2 LEARN IT Types of Dinosaurs Who studies Dinosaurs? KNOW IT USE IT T - Rex A palaeontologist. Theme - Dinosaurs Tyrannosaurus rex is often abbreviated to T-Rex. What YOU need to know! Tyrannosaurus rex walked on two legs, balancing its huge head with a long and heavy tail. The largest tooth of any carnivorous dinosaur found to this date is that of a T-Rex. It is estimated to have been around 30cm (12in) long when Key questions: including the root. What does the word dinosaur mean? Stegosaurus The word dinosaur comes from the Greek They are herbivores language and means ‘terrible lizard’. (plant eaters). Fantastic website all about Dinosaurs - http:// Stegosaurus fossils www.sciencekids.co.nz/sciencefacts/dinosaurs.html Who created the word and the meaning? have been found in An English paleontologist Sir Richard Owen western North America in 1842 and it was meant to refer to and more recently in Portugal, indicating that they Dinosaurs impressive size rather than their Fabulous Facts lived in Europe as well. scary appearance. Rather than being carnivores (meat Although the Stegosaurus body was large, the size eaters), the largest dinosaurs were actual- of their brain was only around the size of a dog’s. The Jurassic Period ly herbivores (plant eaters). Key questions: Birds descended from a type of dinosaurs Sir Richard Owen How long were Dinosaurs on the Earth? known as theropods. Dinosaurs ruled the Earth for over 160 Despite being long extinct, dinosaurs are million years. frequently featured in the media. Jurassic When did dinosaurs walk the Earth? Park was made into a film 1993, the story From the Triassic period around 230 million features cloned dinosaurs brought to life years ago through the Jurassic period and Fabulous Facts until the end of the Cretaceous period Born: 20 July 1804, Lancaster around 65 million years ago. -

A Four-Legged Megalosaurus and Swimming Brontosaurs
Channels: Where Disciplines Meet Volume 2 Number 2 Spring 2018 Article 5 April 2018 A Four-Legged Megalosaurus and Swimming Brontosaurs Jordan C. Oldham Cedarville University, [email protected] Follow this and additional works at: https://digitalcommons.cedarville.edu/channels Part of the Geology Commons, History of Philosophy Commons, Paleontology Commons, and the Philosophy of Science Commons DigitalCommons@Cedarville provides a publication platform for fully open access journals, which means that all articles are available on the Internet to all users immediately upon publication. However, the opinions and sentiments expressed by the authors of articles published in our journals do not necessarily indicate the endorsement or reflect the views of DigitalCommons@Cedarville, the Centennial Library, or Cedarville University and its employees. The authors are solely responsible for the content of their work. Please address questions to [email protected]. Recommended Citation Oldham, Jordan C. (2018) "A Four-Legged Megalosaurus and Swimming Brontosaurs," Channels: Where Disciplines Meet: Vol. 2 : No. 2 , Article 5. DOI: 10.15385/jch.2018.2.2.5 Available at: https://digitalcommons.cedarville.edu/channels/vol2/iss2/5 A Four-Legged Megalosaurus and Swimming Brontosaurs Abstract Thomas Kuhn in his famous work The Structure of Scientific Revolutions laid out the framework for his theory of how science changes. At the advent of dinosaur paleontology fossil hunters like Gideon Mantell discovered some of the first dinosaurs like Iguanodon and Megalosaurus. Through new disciples like Georges Cuvier’s comparative anatomy lead early dinosaur paleontologist to reconstruct them like giant reptiles of absurd proportions. This lead to the formation of a new paradigm that prehistoric animals like dinosaurs existed and eventually went extinct. -
![How Famous and Respected Was Wallace? [Version 3] by Alan Leyin (Thurrock Local History Society, Grays, Essex, UK), October 2014](https://docslib.b-cdn.net/cover/9642/how-famous-and-respected-was-wallace-version-3-by-alan-leyin-thurrock-local-history-society-grays-essex-uk-october-2014-2009642.webp)
How Famous and Respected Was Wallace? [Version 3] by Alan Leyin (Thurrock Local History Society, Grays, Essex, UK), October 2014
How Famous and Respected was Wallace? [Version 3] By Alan Leyin (Thurrock Local History Society, Grays, Essex, UK), October 2014 Having read George Beccaloni’s posting (Wallace Memorial Fund News blog, 25.04.2014) on John van Wyhe’s assertion that it is incorrect to describe Wallace as having been ‘… among the most famous Victorian scientists during his lifetime’; and that the Victorian naturalist ‘… never approached anything like the level of fame or respect’ (van Wyhe 2013, p 172) of many of his contemporaries, I had the urge to rush to my keyboard. Not wishing to duplicate Charles Smith’s response (The Linnean, 2014), to which is referred; nor being able to match either Smith’s or van Wyhe’s command of the Wallace archive, I make a few of observations (some also made elsewhere, on earlier postings) from the point of view of a non-specialist. Being quite different concepts, there is limited value in addressing issues of ‘fame or respect’ as if they were synonymous. One can, of course, accrue respect without being famous; or, indeed, become famous for being notoriously disrespected. To unpick some of the issues… On Wednesday 26th April 1882, a host of the nation’s worthies and international dignitaries gathered at Westminster Abbey to pay their final respects to Charles Darwin. Among those invited were the pre- eminent scientists of the Victorian era; all of whom would have watched the pall-bearers – three members of which had been drawn from their own distinguished circle – carry the unpolished oak coffin from its temporary sanctum in the Chapter House to its place in front of the Communion rails of the Abbey. -

Sir Richard Owen, 1804-1892
Sir Richard Owen, 1804-1892. Shortly before the publication of Darwin's Origin of Species Sir Richard Owen was, by far, the most eminent biologist in Britain, mentioned in the same breath as Newton and Faraday. By this time he had received three gold medals from the Royal Society, of which he had been a member for many years, he was an honorary member of most European Acadamies, had been knighted, had received honorary doctorates from Oxford, Cambridge, Edinburgh and many Continental universities, had a Grace and Favour house in Regents Park and a Civil list pension. Altogether he had received the unprecedented total of over one hundred honours. Yet today he is almost forgotten and his only memorial in his home town of Lancaster is a small figure on the bronze plaque of scientists on the Victoria memorial in Dalton Square. Apart from his remarkable innate talent as a dissector, writer and illustrator, Owen's success was due to several lucky chances. His medical career started inauspiciously. The first doctor he was apprenticed ro quickly died, the second soon joined the Navy while the third was an idle drunk. Curiously it was the lasr who gave Owen his first chance. As prison doctor it was his duty to carry out post mortems on the not inconsiderable number of prisoners who died in goal. He soon left this task to the young Owen. Consequently, at a time when most medical students were lucky to have a part share in a cadaver, it was the time of Burke and Hare, Owen found himself with an almost limitless supply on which to practise.