Structure and Molecular Dynamics Simulations of Protein Tyrosine Phosphatase Non-Receptor 12 Provide Insights Into the Catalytic Mechanism of the Enzyme
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deciphering the Functions of Ets2, Pten and P53 in Stromal Fibroblasts in Multiple
Deciphering the Functions of Ets2, Pten and p53 in Stromal Fibroblasts in Multiple Breast Cancer Models DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Julie Wallace Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2013 Dissertation Committee: Michael C. Ostrowski, PhD, Advisor Gustavo Leone, PhD Denis Guttridge, PhD Dawn Chandler, PhD Copyright by Julie Wallace 2013 Abstract Breast cancer is the second most common cancer in American women, and is also the second leading cause of cancer death in women. It is estimated that nearly a quarter of a million new cases of invasive breast cancer will be diagnosed in women in the United States this year, and approximately 40,000 of these women will die from breast cancer. Although death rates have been on the decline for the past decade, there is still much we need to learn about this disease to improve prevention, detection and treatment strategies. The majority of early studies have focused on the malignant tumor cells themselves, and much has been learned concerning mutations, amplifications and other genetic and epigenetic alterations of these cells. However more recent work has acknowledged the strong influence of tumor stroma on the initiation, progression and recurrence of cancer. Under normal conditions this stroma has been shown to have protective effects against tumorigenesis, however the transformation of tumor cells manipulates this surrounding environment to actually promote malignancy. Fibroblasts in particular make up a significant portion of this stroma, and have been shown to impact various aspects of tumor cell biology. -

2Oc3 Lichtarge Lab 2006
Pages 1–5 2oc3 Evolutionary trace report by report maker September 25, 2008 4.3.3 DSSP 5 4.3.4 HSSP 5 4.3.5 LaTex 5 4.3.6 Muscle 5 4.3.7 Pymol 5 4.4 Note about ET Viewer 5 4.5 Citing this work 5 4.6 About report maker 5 4.7 Attachments 5 1 INTRODUCTION From the original Protein Data Bank entry (PDB id 2oc3): Title: Crystal structure of the catalytic domain of human protein tyrosine phosphatase non-receptor type 18 Compound: Mol id: 1; molecule: tyrosine-protein phosphatase non- receptor type 18; chain: a; synonym: brain-derived phosphatase; ec: CONTENTS 3.1.3.48; engineered: yes Organism, scientific name: 1 Introduction 1 Homo Sapiens; 2oc3 contains a single unique chain 2oc3A (279 residues long). 2 Chain 2oc3A 1 2.1 Q53P42 overview 1 2.2 Multiple sequence alignment for 2oc3A 1 2.3 Residue ranking in 2oc3A 1 2.4 Top ranking residues in 2oc3A and their position on the structure 1 2 CHAIN 2OC3A 2.4.1 Clustering of residues at 25% coverage. 2 2.1 Q53P42 overview 2.4.2 Possible novel functional surfaces at 25% From SwissProt, id Q53P42, 96% identical to 2oc3A: coverage. 2 Description: Hypothetical protein PTPN18. Organism, scientific name: Homo sapiens (Human). 3 Notes on using trace results 3 Taxonomy: Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; 3.1 Coverage 3 Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Primates; 3.2 Known substitutions 3 Catarrhini; Hominidae; Homo. 3.3 Surface 4 3.4 Number of contacts 4 3.5 Annotation 4 3.6 Mutation suggestions 4 2.2 Multiple sequence alignment for 2oc3A 4 Appendix 4 For the chain 2oc3A, the alignment 2oc3A.msf (attached) with 49 4.1 File formats 4 sequences was used. -

Protein Tyrosine Phosphatase Sigma (Ptpσ) Targets Apical Junction Complex Proteins in the Intestine and Modulates
Protein tyrosine phosphatase sigma (PTPσ) targets apical junction complex proteins in the intestine and modulates epithelial permeability. by Ryan Murchie A thesis submitted in conformity with the requirements for the degree of Master of Science Department of Biochemistry University of Toronto © Copyright by Ryan Murchie 2013 Protein tyrosine phosphatase sigma (PTPσ) targets apical junction complex proteins in the intestine and modulates epithelial permeability. Ryan Murchie Master of Science Department of Biochemistry University of Toronto 2012 Abstract Protein tyrosine phosphatase sigma (PTPσ), encoded by PTPRS, was shown previously by us to contain SNP polymorphisms that can confer susceptibility to inflammatory bowel disease (IBD). PTPσ(-/-) mice exhibit an IBD-like phenotype and show increased susceptibility to acute models of murine colitis. The function of PTPσ in the intestine is uncharacterized. Here, I show an intestinal epithelial barrier defect in the PTPσ(-/-) mouse, demonstrated by a decrease in trans-epithelial resistance and a leaky intestinal epithelium that was determined by in vivo tracer analysis. We identified several putative PTPσ intestinal substrates; among these were several proteins that form and regulate the apical junction complex, including ezrin. My results show that ezrin binds to and undergoes tyrosine de-phosphorylation by PTPσ in vitro, suggesting it is a direct substrate for this PTP. The results suggest a role for PTPσ as a positive regulator of epithelial barrier integrity in the intestine. The proteins identified in the screen, including ezrin, suggest that PTPσ may modulate epithelial cell adhesion through the targeting of AJC-associated proteins, a process impaired in IBD. ii Acknowledgements First and foremost, I would like to thank my supervisors Dr. -

PTPN18 Rabbit Pab
Leader in Biomolecular Solutions for Life Science PTPN18 Rabbit pAb Catalog No.: A8487 Basic Information Background Catalog No. The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) A8487 family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, the mitotic cycle, and oncogenic Observed MW transformation. This PTP contains a PEST motif, which often serves as a protein-protein 60kDa interaction domain, and may be related to protein intracellular half-live. This protein can differentially dephosphorylate autophosphorylated tyrosine kinases that are Calculated MW overexpressed in tumor tissues, and it appears to regulate HER2, a member of the 38kDa/50kDa epidermal growth factor receptor family of receptor tyrosine kinases. Two transcript variants encoding different isoforms have been found for this gene. Category Primary antibody Applications WB Cross-Reactivity Human Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 26469 Q99952 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 1-210 of human PTPN18 (NP_001135842.1). Synonyms PTPN18;BDP1;PTP-HSCF Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using PTPN18 antibody (A8487) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. Detection: ECL Basic Kit (RM00020). -

Live-Cell Imaging Rnai Screen Identifies PP2A–B55α and Importin-Β1 As Key Mitotic Exit Regulators in Human Cells
LETTERS Live-cell imaging RNAi screen identifies PP2A–B55α and importin-β1 as key mitotic exit regulators in human cells Michael H. A. Schmitz1,2,3, Michael Held1,2, Veerle Janssens4, James R. A. Hutchins5, Otto Hudecz6, Elitsa Ivanova4, Jozef Goris4, Laura Trinkle-Mulcahy7, Angus I. Lamond8, Ina Poser9, Anthony A. Hyman9, Karl Mechtler5,6, Jan-Michael Peters5 and Daniel W. Gerlich1,2,10 When vertebrate cells exit mitosis various cellular structures can contribute to Cdk1 substrate dephosphorylation during vertebrate are re-organized to build functional interphase cells1. This mitotic exit, whereas Ca2+-triggered mitotic exit in cytostatic-factor- depends on Cdk1 (cyclin dependent kinase 1) inactivation arrested egg extracts depends on calcineurin12,13. Early genetic studies in and subsequent dephosphorylation of its substrates2–4. Drosophila melanogaster 14,15 and Aspergillus nidulans16 reported defects Members of the protein phosphatase 1 and 2A (PP1 and in late mitosis of PP1 and PP2A mutants. However, the assays used in PP2A) families can dephosphorylate Cdk1 substrates in these studies were not specific for mitotic exit because they scored pro- biochemical extracts during mitotic exit5,6, but how this relates metaphase arrest or anaphase chromosome bridges, which can result to postmitotic reassembly of interphase structures in intact from defects in early mitosis. cells is not known. Here, we use a live-cell imaging assay and Intracellular targeting of Ser/Thr phosphatase complexes to specific RNAi knockdown to screen a genome-wide library of protein substrates is mediated by a diverse range of regulatory and targeting phosphatases for mitotic exit functions in human cells. We subunits that associate with a small group of catalytic subunits3,4,17. -

Expression Profile of Tyrosine Phosphatases in HER2 Breast
Cellular Oncology 32 (2010) 361–372 361 DOI 10.3233/CLO-2010-0520 IOS Press Expression profile of tyrosine phosphatases in HER2 breast cancer cells and tumors Maria Antonietta Lucci a, Rosaria Orlandi b, Tiziana Triulzi b, Elda Tagliabue b, Andrea Balsari c and Emma Villa-Moruzzi a,∗ a Department of Experimental Pathology, University of Pisa, Pisa, Italy b Molecular Biology Unit, Department of Experimental Oncology, Istituto Nazionale Tumori, Milan, Italy c Department of Human Morphology and Biomedical Sciences, University of Milan, Milan, Italy Abstract. Background: HER2-overexpression promotes malignancy by modulating signalling molecules, which include PTPs/DSPs (protein tyrosine and dual-specificity phosphatases). Our aim was to identify PTPs/DSPs displaying HER2-associated expression alterations. Methods: HER2 activity was modulated in MDA-MB-453 cells and PTPs/DSPs expression was analysed with a DNA oligoar- ray, by RT-PCR and immunoblotting. Two public breast tumor datasets were analysed to identify PTPs/DSPs differentially ex- pressed in HER2-positive tumors. Results: In cells (1) HER2-inhibition up-regulated 4 PTPs (PTPRA, PTPRK, PTPN11, PTPN18) and 11 DSPs (7 MKPs [MAP Kinase Phosphatases], 2 PTP4, 2 MTMRs [Myotubularin related phosphatases]) and down-regulated 7 DSPs (2 MKPs, 2 MTMRs, CDKN3, PTEN, CDC25C); (2) HER2-activation with EGF affected 10 DSPs (5 MKPs, 2 MTMRs, PTP4A1, CDKN3, CDC25B) and PTPN13; 8 DSPs were found in both groups. Furthermore, 7 PTPs/DSPs displayed also altered protein level. Analysis of 2 breast cancer datasets identified 6 differentially expressed DSPs: DUSP6, strongly up-regulated in both datasets; DUSP10 and CDC25B, up-regulated; PTP4A2, CDC14A and MTMR11 down-regulated in one dataset. -

Phosphatases Page 1
Phosphatases esiRNA ID Gene Name Gene Description Ensembl ID HU-05948-1 ACP1 acid phosphatase 1, soluble ENSG00000143727 HU-01870-1 ACP2 acid phosphatase 2, lysosomal ENSG00000134575 HU-05292-1 ACP5 acid phosphatase 5, tartrate resistant ENSG00000102575 HU-02655-1 ACP6 acid phosphatase 6, lysophosphatidic ENSG00000162836 HU-13465-1 ACPL2 acid phosphatase-like 2 ENSG00000155893 HU-06716-1 ACPP acid phosphatase, prostate ENSG00000014257 HU-15218-1 ACPT acid phosphatase, testicular ENSG00000142513 HU-09496-1 ACYP1 acylphosphatase 1, erythrocyte (common) type ENSG00000119640 HU-04746-1 ALPL alkaline phosphatase, liver ENSG00000162551 HU-14729-1 ALPP alkaline phosphatase, placental ENSG00000163283 HU-14729-1 ALPP alkaline phosphatase, placental ENSG00000163283 HU-14729-1 ALPPL2 alkaline phosphatase, placental-like 2 ENSG00000163286 HU-07767-1 BPGM 2,3-bisphosphoglycerate mutase ENSG00000172331 HU-06476-1 BPNT1 3'(2'), 5'-bisphosphate nucleotidase 1 ENSG00000162813 HU-09086-1 CANT1 calcium activated nucleotidase 1 ENSG00000171302 HU-03115-1 CCDC155 coiled-coil domain containing 155 ENSG00000161609 HU-09022-1 CDC14A CDC14 cell division cycle 14 homolog A (S. cerevisiae) ENSG00000079335 HU-11533-1 CDC14B CDC14 cell division cycle 14 homolog B (S. cerevisiae) ENSG00000081377 HU-06323-1 CDC25A cell division cycle 25 homolog A (S. pombe) ENSG00000164045 HU-07288-1 CDC25B cell division cycle 25 homolog B (S. pombe) ENSG00000101224 HU-06033-1 CDKN3 cyclin-dependent kinase inhibitor 3 ENSG00000100526 HU-02274-1 CTDSP1 CTD (carboxy-terminal domain, -

The Biological Age of a Bloodstain Donor Author(S): Jack Ballantyne, Ph.D
The author(s) shown below used Federal funding provided by the U.S. Department of Justice to prepare the following resource: Document Title: The Biological Age of a Bloodstain Donor Author(s): Jack Ballantyne, Ph.D. Document Number: 251894 Date Received: July 2018 Award Number: 2009-DN-BX-K179 This resource has not been published by the U.S. Department of Justice. This resource is being made publically available through the Office of Justice Programs’ National Criminal Justice Reference Service. Opinions or points of view expressed are those of the author(s) and do not necessarily reflect the official position or policies of the U.S. Department of Justice. National Center for Forensic Science University of Central Florida P.O. Box 162367 · Orlando, FL 32826 Phone: 407.823.4041 Fax: 407.823.4042 Web site: http://www.ncfs.org/ Biological Evidence _________________________________________________________________________________________________________ The Biological Age of a Bloodstain Donor FINAL REPORT May 27, 2014 Department of Justice, National Institute of justice Award Number: 2009-DN-BX-K179 (1 October 2009 – 31 May 2014) _________________________________________________________________________________________________________ Principal Investigator: Jack Ballantyne, PhD Professor Department of Chemistry Associate Director for Research National Center for Forensic Science P.O. Box 162367 Orlando, FL 32816-2366 Phone: (407) 823 4440 Fax: (407) 823 4042 e-mail: [email protected] 1 This resource was prepared by the author(s) using Federal funds provided by the U.S. Department of Justice. Opinions or points of view expressed are those of the author(s) and do not necessarily reflect the official position or policies of the U.S. -

PTPN18 Conjugated Antibody
Product Datasheet PTPN18 Conjugated Antibody Catalog No: #C31565 Package Size: #C31565-AF350 100ul #C31565-AF405 100ul #C31565-AF488 100ul Orders: [email protected] Support: [email protected] #C31565-AF555 100ul #C31565-AF594 100ul #C31565-AF647 100ul #C31565-AF680 100ul #C31565-AF750 100ul #C31565-Biotin 100ul Description Product Name PTPN18 Conjugated Antibody Host Species Rabbit Clonality Polyclonal Isotype IgG Purification Affinity purification Applications most applications Species Reactivity Hu Immunogen Description Recombinant fusion protein of human PTPN18 (NP_001135842.1). Conjugates Biotin AF350 AF405 AF488 AF555 AF594 AF647 AF680 AF750 Other Names PTPN18; BDP1; PTP-HSCF; tyrosine-protein phosphatase non-receptor type 18 Accession No. Swiss-Prot#:Q99952NCBI Gene ID:26469 Calculated MW 60kDa Formulation 0.01M Sodium Phosphate, 0.25M NaCl, pH 7.6, 5mg/ml Bovine Serum Albumin, 0.02% Sodium Azide Storage Store at 4°C in dark for 6 months Application Details Suggested Dilution: AF350 conjugated: most applications: 1: 50 - 1: 250 AF405 conjugated: most applications: 1: 50 - 1: 250 AF488 conjugated: most applications: 1: 50 - 1: 250 AF555 conjugated: most applications: 1: 50 - 1: 250 AF594 conjugated: most applications: 1: 50 - 1: 250 AF647 conjugated: most applications: 1: 50 - 1: 250 AF680 conjugated: most applications: 1: 50 - 1: 250 AF750 conjugated: most applications: 1: 50 - 1: 250 Biotin conjugated: working with enzyme-conjugated streptavidin, most applications: 1: 50 - 1: 1,000 Background The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, the mitotic cycle, and oncogenic transformation. -

Protein Tyrosine Phosphatases in Health and Disease Wiljan J
REVIEW ARTICLE Protein tyrosine phosphatases in health and disease Wiljan J. A. J. Hendriks1, Ari Elson2, Sheila Harroch3, Rafael Pulido4, Andrew Stoker5 and Jeroen den Hertog6,7 1 Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands 2 Department of Molecular Genetics, The Weizmann Institute of Science, Rehovot, Israel 3 Department of Neuroscience, Institut Pasteur, Paris, France 4 Centro de Investigacio´ nPrı´ncipe Felipe, Valencia, Spain 5 Neural Development Unit, Institute of Child Health, University College London, UK 6 Hubrecht Institute, KNAW & University Medical Center Utrecht, The Netherlands 7 Institute of Biology Leiden, Leiden University, The Netherlands Keywords Protein tyrosine phosphatases (PTPs) represent a super-family of enzymes bone morphogenesis; hereditary disease; that play essential roles in normal development and physiology. In this neuronal development; post-translational review, we will discuss the PTPs that have a causative role in hereditary modification; protein tyrosine phosphatase; diseases in humans. In addition, recent progress in the development and synaptogenesis analysis of animal models expressing mutant PTPs will be presented. The Correspondence impact of PTP signaling on health and disease will be exemplified for the J. den Hertog, Hubrecht Institute, fields of bone development, synaptogenesis and central nervous system dis- Uppsalalaan 8, 3584 CT Utrecht, eases. Collectively, research on PTPs since the late 1980’s yielded the The Netherlands cogent view that development of PTP-directed -
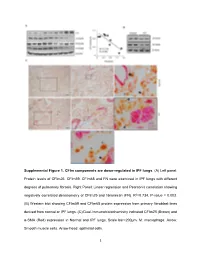
1 Supplemental Figure 1. Cfim Components Are Down-Regulated In
Supplemental Figure 1. CFIm components are down-regulated in IPF lungs. (A) Left panel: Protein levels of CFIm25, CFIm59, CFIm68 and FN were examined in IPF lungs with different degrees of pulmonary fibrosis. Right Panel: Linear regression and Pearson’s correlation showing negatively correlated densitometry of CFIm25 and fibronectin (FN). R2=0.734, P-value = 0.003. (B) Western blot showing CFIm59 and CFIm68 protein expression from primary fibroblast lines derived from normal or IPF lungs. (C) Dual-Immunohistochemistry indicated CFIm25 (Brown) and α-SMA (Red) expression in Normal and IPF lungs. Scale bar=200µm. M: macrophage. Arrow: Smooth muscle cells. Arrow head: epithelial cells. 1 Supplemental Figure 2. CFIm components are down-regulated in an animal model with pulmonary fibrosis. (A) Western blot analysis of the protein expression of CFIm59 and CFIm68 in whole lung lysates at day 33 after PBS or bleomycin administration. (B) Immunohistochemistry was carried out to determine CFIm25 (brown) and α-SMA (pink) localization in lungs from mice exposed to PBS or bleomycin for 33 days. Arrow: CFIm25 positive cells. Arrow head: α-SMA positive but CFIm25 negative cells. Scale bar=100 µm. (C) Western blot was used to determine CFIm59, CFIm68 and β-actin protein levels in primary fibroblasts isolated from day 33 PBS or bleomycin-injected mouse lungs. 2 Supplemental Figure 3. Components in TGFβ and Wnt pathways have 3’UTR shortening after CFIm25 knockdown. KEGG pathway shows genes having 3’UTR shortening (green) or lengthening (red) in TGFβ (A) and Wnt pathways (B) in CFIm25 knockdown CCD8-Lu cells compared to controls. -

Inhibitory Enzymes SHIP1 and Csk Autoinflammatory Disease, Interacts
PSTPIP2, a Protein Associated with Autoinflammatory Disease, Interacts with Inhibitory Enzymes SHIP1 and Csk This information is current as Ales Drobek, Jarmila Kralova, Tereza Skopcova, Marketa of September 28, 2021. Kucova, Petr Novák, Pavla Angelisová, Pavel Otahal, Meritxell Alberich-Jorda and Tomas Brdicka J Immunol 2015; 195:3416-3426; Prepublished online 24 August 2015; doi: 10.4049/jimmunol.1401494 Downloaded from http://www.jimmunol.org/content/195/7/3416 Supplementary http://www.jimmunol.org/content/suppl/2015/08/21/jimmunol.140149 Material 4.DCSupplemental http://www.jimmunol.org/ References This article cites 58 articles, 29 of which you can access for free at: http://www.jimmunol.org/content/195/7/3416.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 28, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2015 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology PSTPIP2, a Protein Associated with Autoinflammatory Disease, Interacts with Inhibitory Enzymes SHIP1 and Csk Ales Drobek,* Jarmila Kralova,* Tereza Skopcova,* Marketa Kucova,* Petr Nova´k,† Pavla Angelisova´,‡ Pavel Otahal,‡ Meritxell Alberich-Jorda,x and Tomas Brdicka* Mutations in the adaptor protein PSTPIP2 are the cause of the autoinflammatory disease chronic multifocal osteomyelitis in mice.